
https://www.ed.ac.uk/roslin/highlanderlab













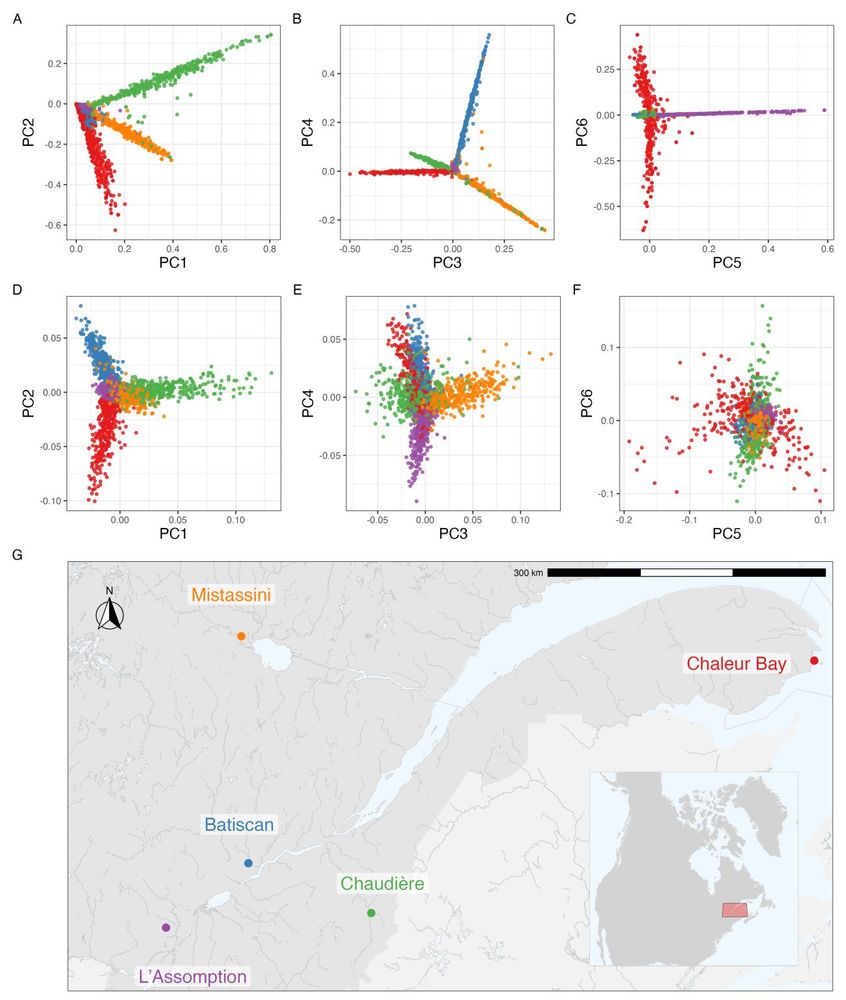

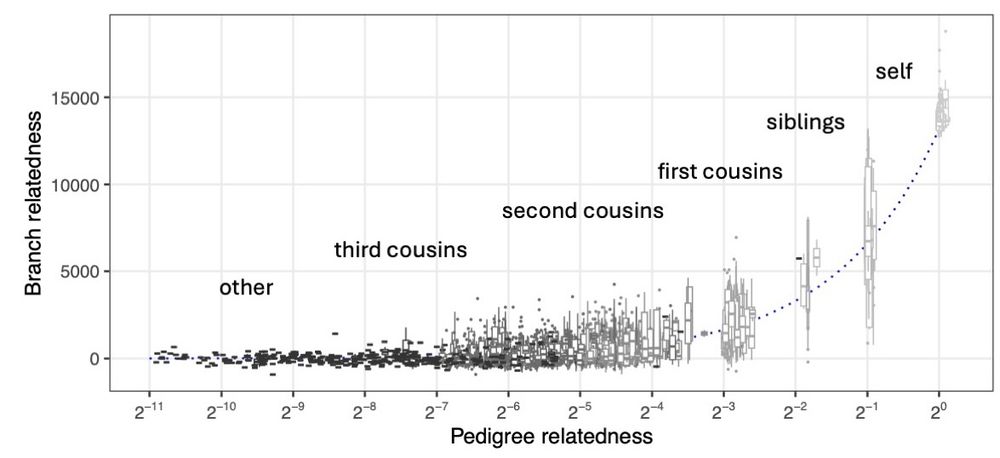
Daniel Tolhurst's abstract was selected for a talk: Disentangling Non-Crossover and Crossover Genotype by Environment Interaction for Selection

Daniel Tolhurst's abstract was selected for a talk: Disentangling Non-Crossover and Crossover Genotype by Environment Interaction for Selection
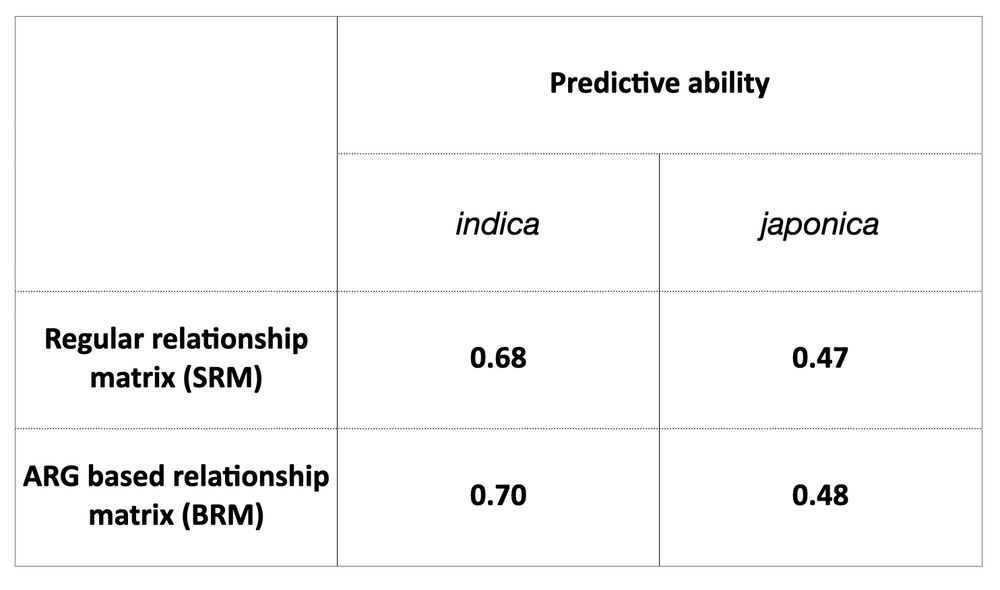
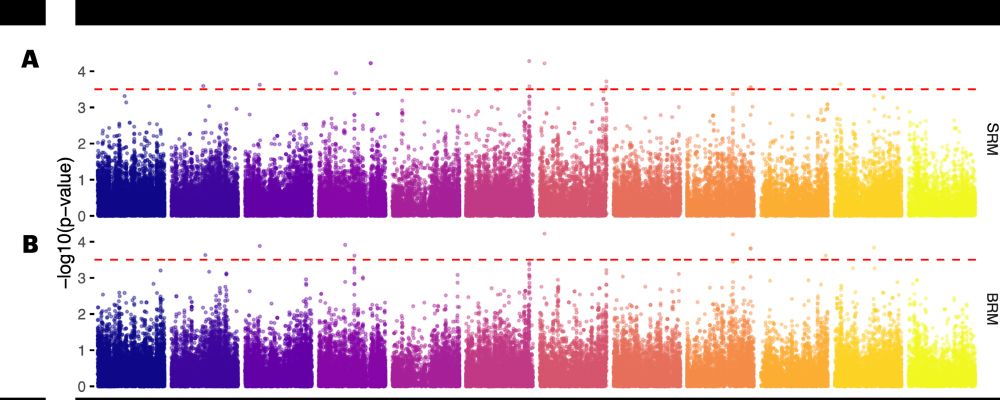
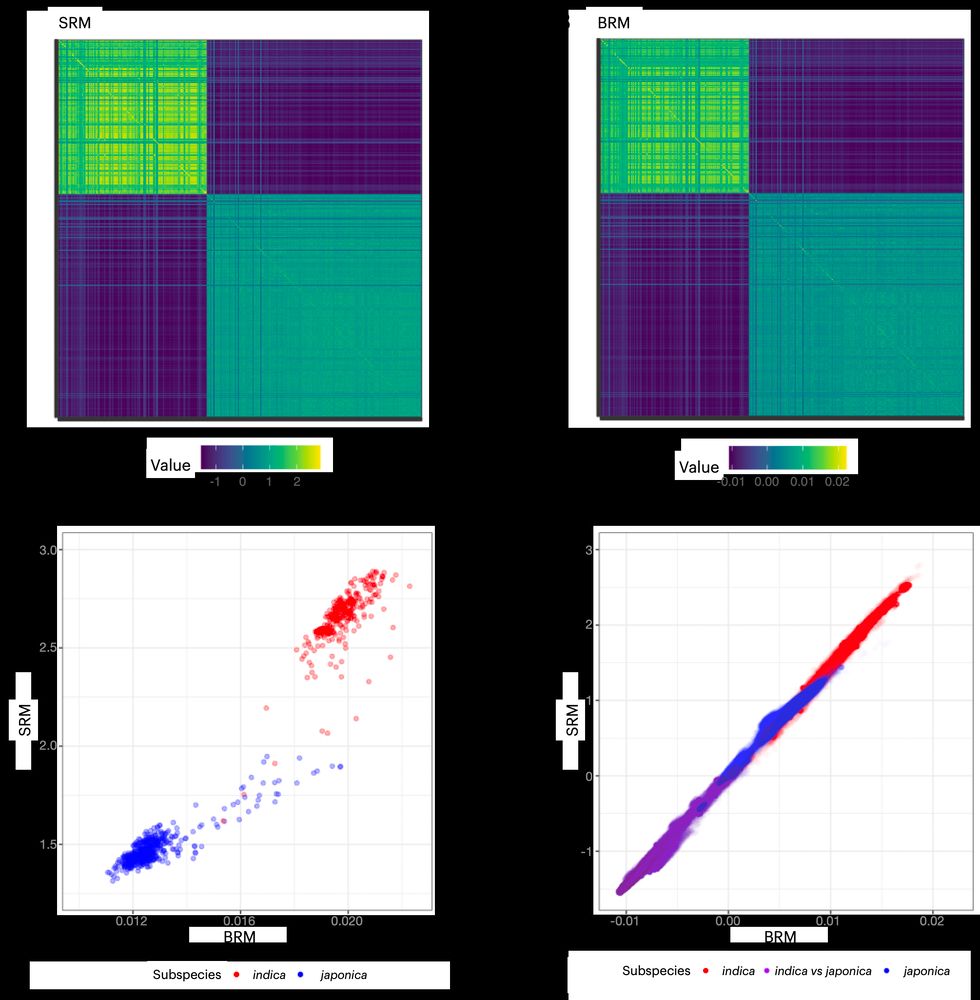
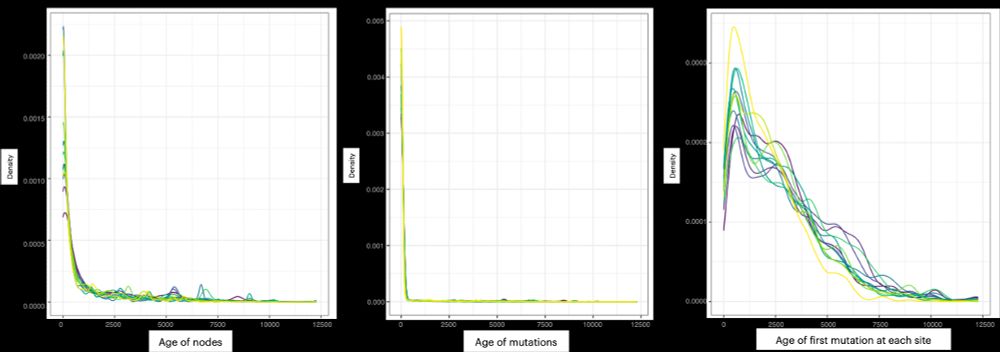
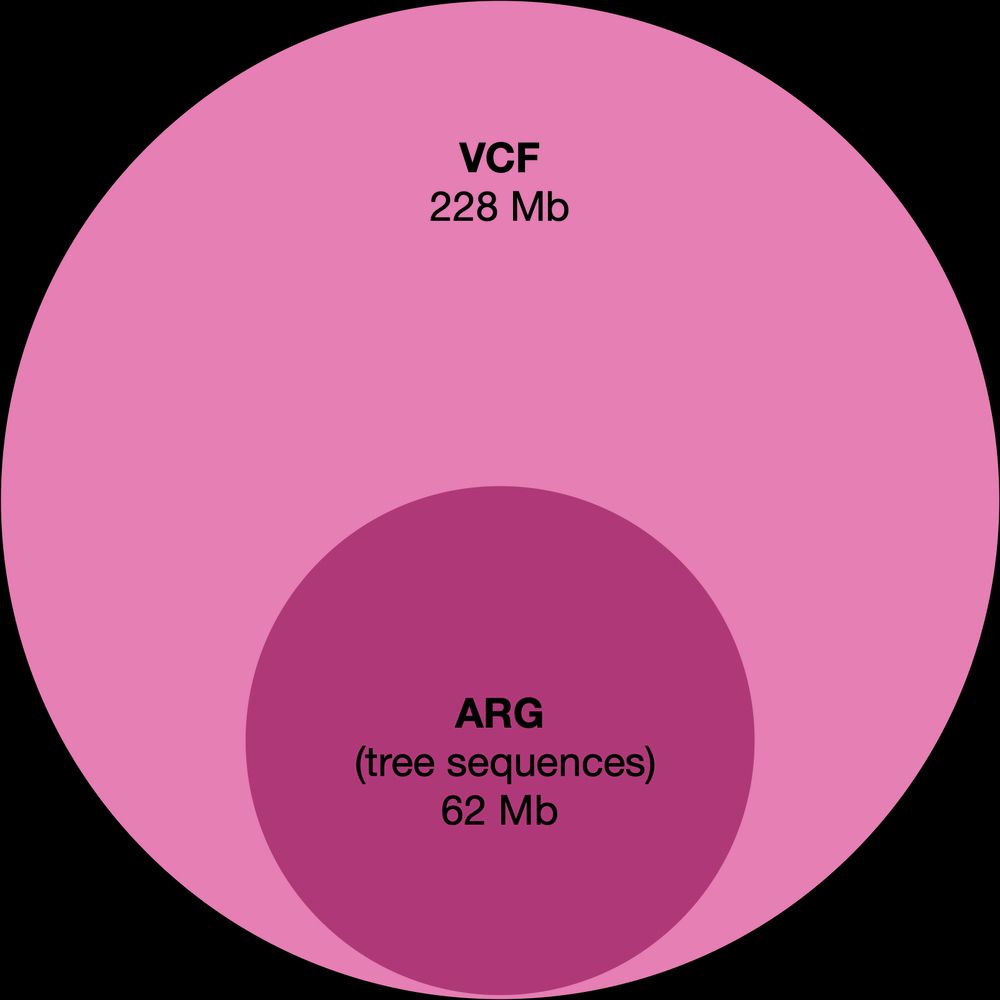
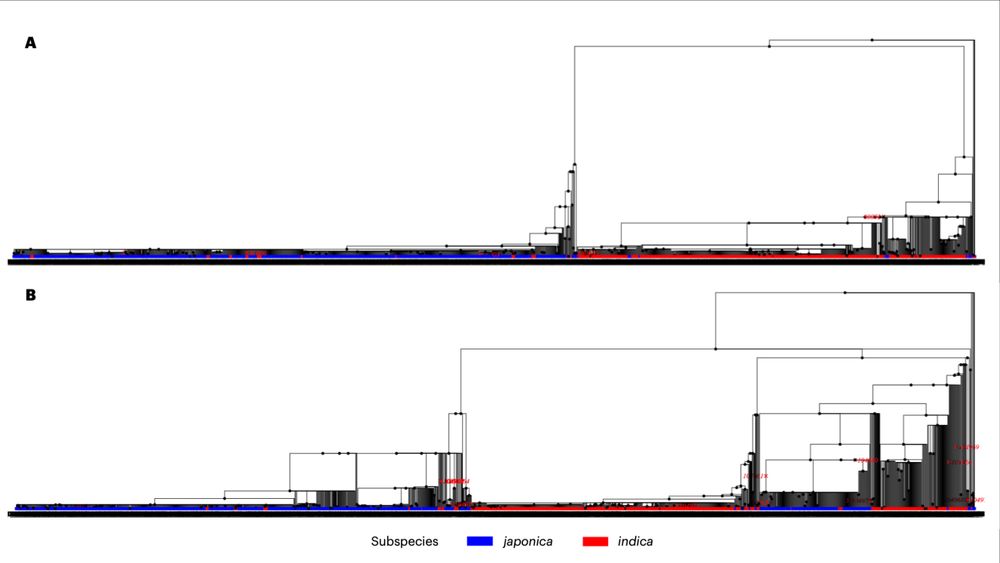



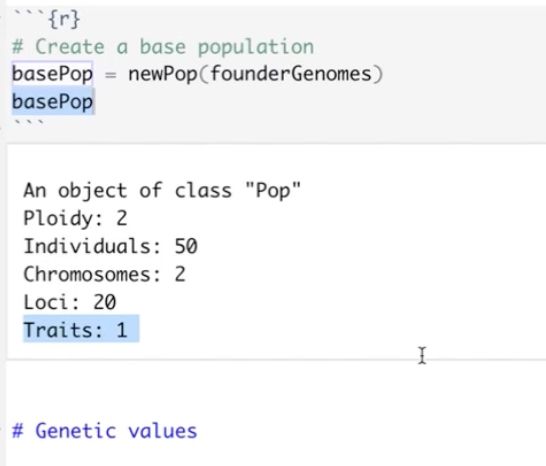
It’s time for trait simulation - one of the key reasons we developed AlphaSimR - to study the principles of quantitative genetics and selective breeding. Here, we show the outline of such a simulation, ...




It’s time for trait simulation - one of the key reasons we developed AlphaSimR - to study the principles of quantitative genetics and selective breeding. Here, we show the outline of such a simulation, ...







