
I am very excited to finally share what has been the main focus of my PhD for the past almost 3 years! It is about viral dark matter and a powerful tool we built to shed light on it. 🧬💡
Continue reading (🧵)

I am very excited to finally share what has been the main focus of my PhD for the past almost 3 years! It is about viral dark matter and a powerful tool we built to shed light on it. 🧬💡
Continue reading (🧵)
Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8

Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8
A bile acid-bound structure of toxin TcdB revealed the mechanism of inhibition and guided the design of a synthetic bile acid that alleviated C. difficile infection in mice.
#MicroSky 🦠💊
www.nature.com/articles/s41...

A bile acid-bound structure of toxin TcdB revealed the mechanism of inhibition and guided the design of a synthetic bile acid that alleviated C. difficile infection in mice.
#MicroSky 🦠💊
www.nature.com/articles/s41...
academic.oup.com/nar/article/... 1/5

academic.oup.com/nar/article/... 1/5
Predicting host- #phage interactions from genomics.
www.biorxiv.org/content/10.1...

Predicting host- #phage interactions from genomics.
www.biorxiv.org/content/10.1...
www.grc.org/microbiome-e...
www.grc.org/microbiome-e...
Thanks to wonderful coauthors/collaborators/friends, the whole @doudna-lab.bsky.social and everyone at @innovativegenomics.bsky.social

Thanks to wonderful coauthors/collaborators/friends, the whole @doudna-lab.bsky.social and everyone at @innovativegenomics.bsky.social
Review article published in @natrevmicro.nature.com with @bupbuse.bsky.social, Andriko von Kügelgen and @vikramalva.bsky.social.
S-layers are everywhere!
www.nature.com/articles/s41...

Review article published in @natrevmicro.nature.com with @bupbuse.bsky.social, Andriko von Kügelgen and @vikramalva.bsky.social.
S-layers are everywhere!
www.nature.com/articles/s41...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
www.pnas.org/doi/10.1073/...
Sadly, the Editors at PNAS rejected our initial introduction, which was a David Attenborough style voice over of the microbial Serengeti (included below)

www.pnas.org/doi/10.1073/...
Sadly, the Editors at PNAS rejected our initial introduction, which was a David Attenborough style voice over of the microbial Serengeti (included below)
We present a foundational genomic resource of human gut microbiome viruses. It delivers high-quality, deeply curated data spanning taxonomy, predicted hosts, structures, and functions, providing a reference for gut virome research. (1/8)
www.biorxiv.org/content/10.1...

We present a foundational genomic resource of human gut microbiome viruses. It delivers high-quality, deeply curated data spanning taxonomy, predicted hosts, structures, and functions, providing a reference for gut virome research. (1/8)
www.biorxiv.org/content/10.1...
Here’s the story of how we discovered the Metis defense system 👇
www.biorxiv.org/content/10.1...

www.pnas.org/doi/10.1073/...

www.pnas.org/doi/10.1073/...

www.nature.com/articles/s41...
Thanks again to our awesome collaborators @mblokesch.bsky.social and David and co and Mark Szczelkun and @steven-shaw.bsky.social and the DCI Lausanne @fbm-unil.bsky.social
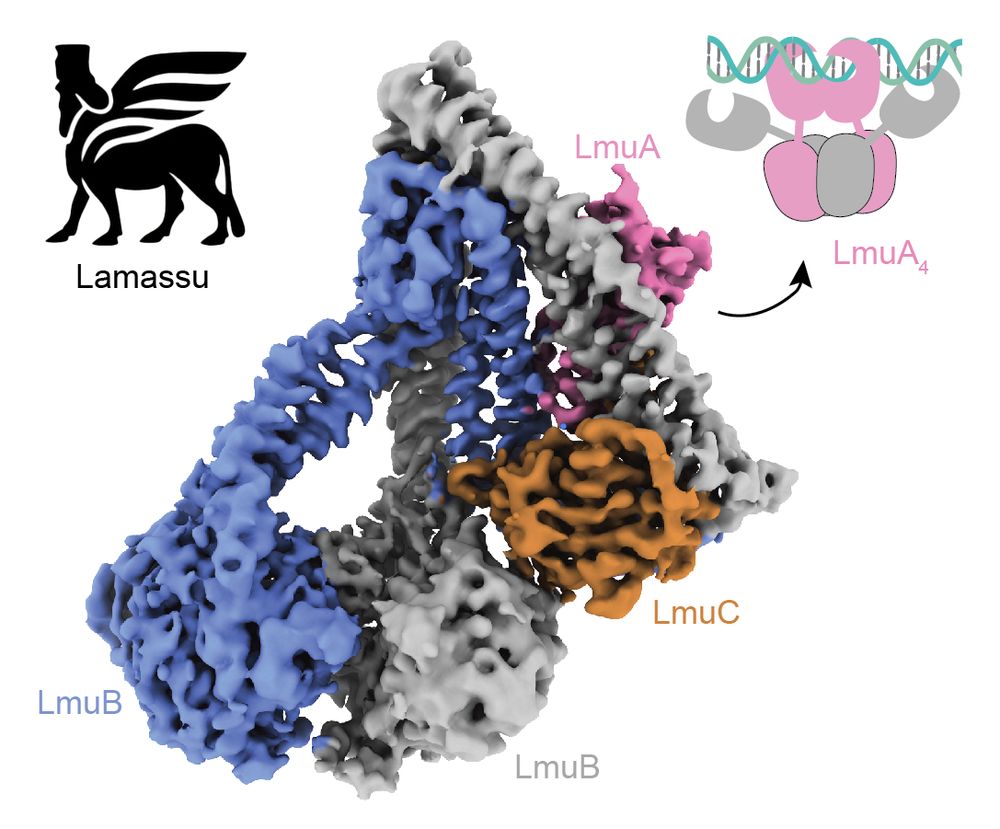
www.nature.com/articles/s41...
Thanks again to our awesome collaborators @mblokesch.bsky.social and David and co and Mark Szczelkun and @steven-shaw.bsky.social and the DCI Lausanne @fbm-unil.bsky.social
Today, we report the discovery of telomerase homologs in a family of antiviral RTs, revealing an unexpected evolutionary origin in bacteria.
www.biorxiv.org/content/10.1...

Today, we report the discovery of telomerase homologs in a family of antiviral RTs, revealing an unexpected evolutionary origin in bacteria.
www.biorxiv.org/content/10.1...
#MicroSky
www.cell.com/molecular-ce...

#MicroSky
www.cell.com/molecular-ce...

We developed new genetic tools & genome-wide libraries for species of the Bacteroidales order; constructed saturated barcoded transposon libraries in key representatives of three genera.
www.biorxiv.org/content/10.1...

We developed new genetic tools & genome-wide libraries for species of the Bacteroidales order; constructed saturated barcoded transposon libraries in key representatives of three genera.
www.biorxiv.org/content/10.1...



