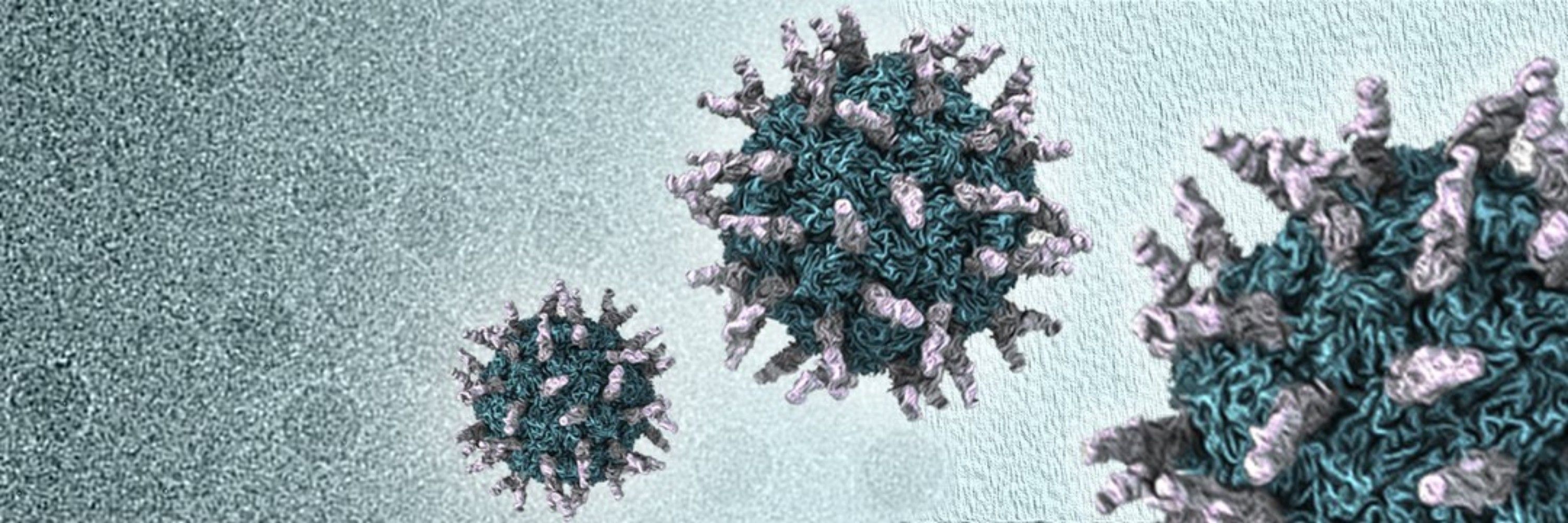
We use #cryoEM and molecular #virology techniques to study the infection mechanisms of +ssRNA viruses.
https://www.uu.nl/medewerkers/DLHurdiss
Views are my own.
They unwind DNA using coordinated, ATP-driven conformational changes across a hexameric ring.
But no equivalent structures are available for SF3 proteins encoded by RNA viruses...
They unwind DNA using coordinated, ATP-driven conformational changes across a hexameric ring.
But no equivalent structures are available for SF3 proteins encoded by RNA viruses...
By combining #cryoEM with #AlphaFold3 modelling, we propose that norovirus NS3 forms a transmembrane RNA translocase.
This could have big implications for our understanding of viral replication & assembly (🧵)
www.biorxiv.org/content/10.1...
By combining #cryoEM with #AlphaFold3 modelling, we propose that norovirus NS3 forms a transmembrane RNA translocase.
This could have big implications for our understanding of viral replication & assembly (🧵)
www.biorxiv.org/content/10.1...
Our latest preprint dives into the highly unusual spikes of marine mammal coronaviruses.
www.biorxiv.org/content/10.1...
This #cryoEM study was led by @viralfusion.bsky.social, with key contributions from an amazing team.
Our latest preprint dives into the highly unusual spikes of marine mammal coronaviruses.
www.biorxiv.org/content/10.1...
This #cryoEM study was led by @viralfusion.bsky.social, with key contributions from an amazing team.
"A coronavirus assembly inhibitor that targets the viral membrane protein"
www.nature.com/articles/s41...
A huge congrats to everyone involved! 🎉
"A coronavirus assembly inhibitor that targets the viral membrane protein"
www.nature.com/articles/s41...
A huge congrats to everyone involved! 🎉

