
https://www.cresslab.bio/

Eliminate your opponents of course.
Recently, my friend @fernpizza.bsky.social showed how plasmids compete intracellularly (check out his paper published in Science today!). With @baym.lol, we now know they can fight.
www.biorxiv.org/content/10.1...

Eliminate your opponents of course.
Recently, my friend @fernpizza.bsky.social showed how plasmids compete intracellularly (check out his paper published in Science today!). With @baym.lol, we now know they can fight.
www.biorxiv.org/content/10.1...
Eliminate your opponents of course.
Recently, my friend @fernpizza.bsky.social showed how plasmids compete intracellularly (check out his paper published in Science today!). With @baym.lol, we now know they can fight.
www.biorxiv.org/content/10.1...

www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
I am very excited to finally share what has been the main focus of my PhD for the past almost 3 years! It is about viral dark matter and a powerful tool we built to shed light on it. 🧬💡
Continue reading (🧵)

I am very excited to finally share what has been the main focus of my PhD for the past almost 3 years! It is about viral dark matter and a powerful tool we built to shed light on it. 🧬💡
Continue reading (🧵)
Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8

Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8


www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
A solid machine learning framework & to predict strain-level phage-host interactions across diverse bacterial genera from genome sequences alone. Avery Noonan from the Arkin Lab led this massive effort
www.biorxiv.org/content/10.1...
A solid machine learning framework & to predict strain-level phage-host interactions across diverse bacterial genera from genome sequences alone. Avery Noonan from the Arkin Lab led this massive effort
www.biorxiv.org/content/10.1...
Predicting host- #phage interactions from genomics.
www.biorxiv.org/content/10.1...

Predicting host- #phage interactions from genomics.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
www.grc.org/microbiome-e...
www.grc.org/microbiome-e...
www.science.org/doi/10.1126/...

www.science.org/doi/10.1126/...
The Dept of Biological and Environmental Science, University of Jyväskylä, is recruiting 1–4 new PhD candidates.
One of the positions is potentially in my lab, focusing on megaphages and their hosts.
Details and apply here 👇
bit.ly/48eqYNB
#phage
The Dept of Biological and Environmental Science, University of Jyväskylä, is recruiting 1–4 new PhD candidates.
One of the positions is potentially in my lab, focusing on megaphages and their hosts.
Details and apply here 👇
bit.ly/48eqYNB
#phage
Meet MetaEdit: a platform for pathway-scale metagenomic editing inside the gut microbiome. science.org/doi/10.1126/...

Meet MetaEdit: a platform for pathway-scale metagenomic editing inside the gut microbiome. science.org/doi/10.1126/...
Three SWBio DTP PhD project opportunities linked to the MultiDefence consortium are now open (1/4)
#PhD #DoctoralTraining #ResearchCarrers #LifeSciences #phagesky #AMR #microsky #PhDPositions
Three SWBio DTP PhD project opportunities linked to the MultiDefence consortium are now open (1/4)
#PhD #DoctoralTraining #ResearchCarrers #LifeSciences #phagesky #AMR #microsky #PhDPositions


Thanks to wonderful coauthors/collaborators/friends, the whole @doudna-lab.bsky.social and everyone at @innovativegenomics.bsky.social

Thanks to wonderful coauthors/collaborators/friends, the whole @doudna-lab.bsky.social and everyone at @innovativegenomics.bsky.social

Thanks to wonderful coauthors/collaborators/friends, the whole @doudna-lab.bsky.social and everyone at @innovativegenomics.bsky.social
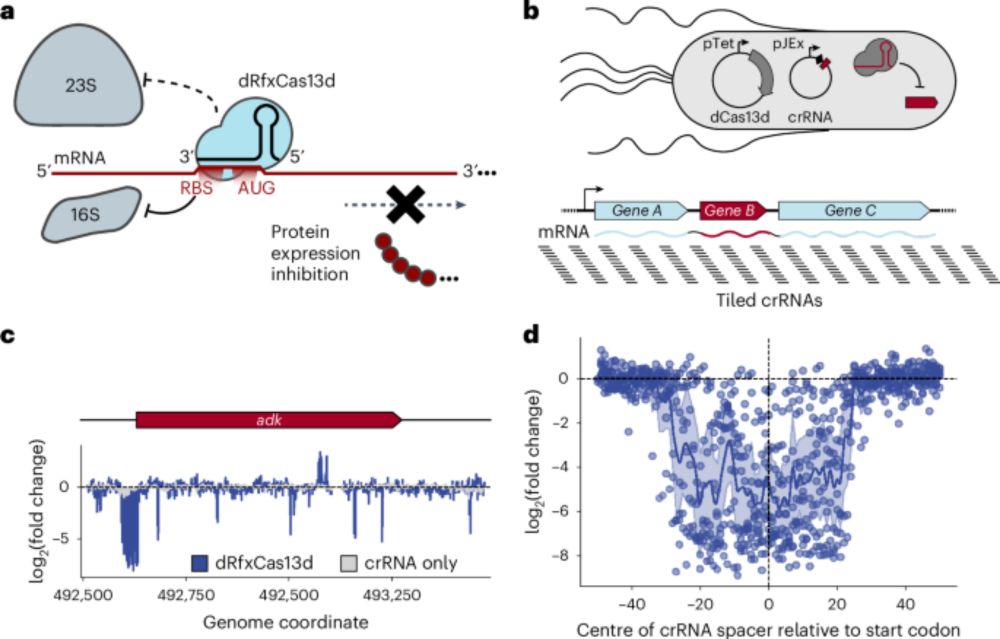
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.
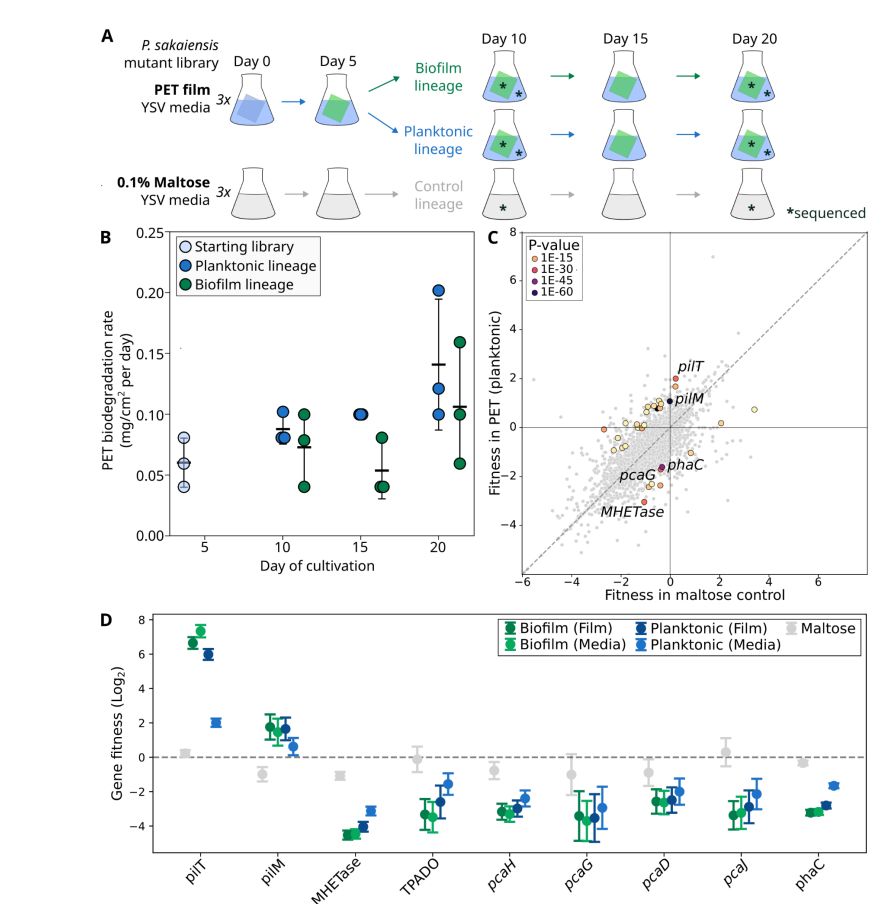
We identified a plasmid vector for the strain and generated a large RB-TnSeq library, screening for genes impacting plastic degradation.

vConTACT3 delivers a unified, scalable, and transparent framework for genome-based virus taxonomy — helping translate big viral data into systematic classification.
🔗 Read the preprint: doi.org/10.1101/2025...
Improvements details below 👇

vConTACT3 delivers a unified, scalable, and transparent framework for genome-based virus taxonomy — helping translate big viral data into systematic classification.
🔗 Read the preprint: doi.org/10.1101/2025...
Improvements details below 👇

