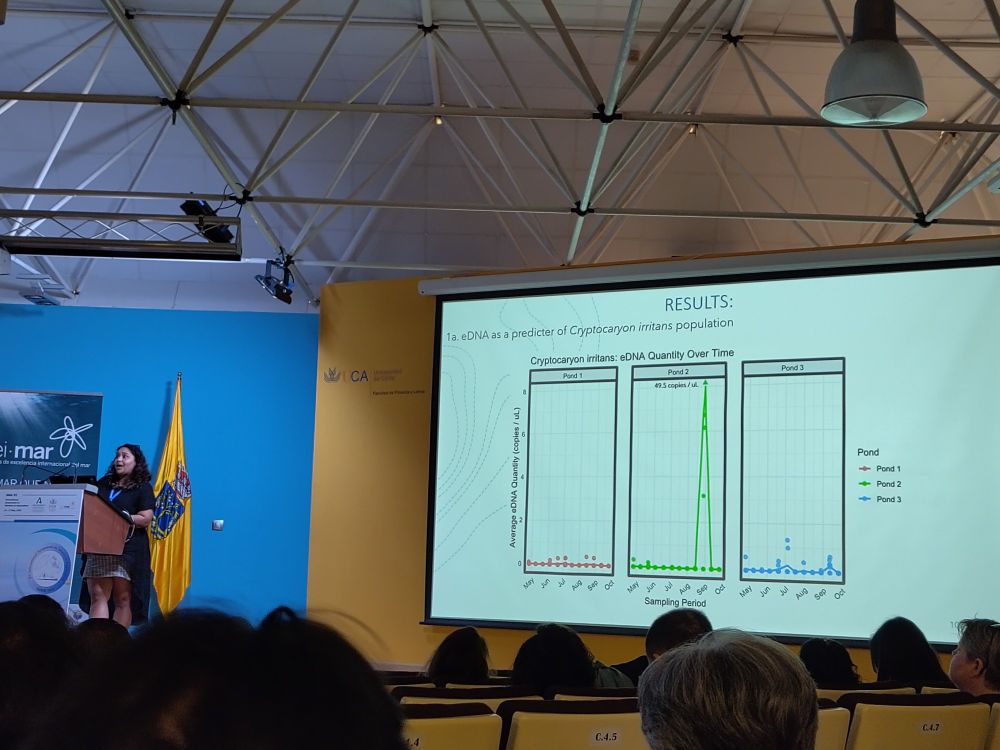
From France, raised in Madagascar and now in Edinburgh!
This work was done within the AquaImpact project (Horizon 2020) and in collaboration with Mowi Genetics using datasets from there breeding programme. 🐟🧬
This work was done within the AquaImpact project (Horizon 2020) and in collaboration with Mowi Genetics using datasets from there breeding programme. 🐟🧬
Only the parents (all traits together or one trait at a time ie: increasing the target population size)
Only the selection candidates (validation dataset)
Parents and selection candidates
Parents and some collaterals (100-1000)
in 3 traits: Growth, resistance to PD and to CMS
Only the parents (all traits together or one trait at a time ie: increasing the target population size)
Only the selection candidates (validation dataset)
Parents and selection candidates
Parents and some collaterals (100-1000)
in 3 traits: Growth, resistance to PD and to CMS
🧪
🧬💻
🧪
🧬💻
Sarah is leaving the Roslin and starting her group in @exeter.ac.uk, she will be missed !
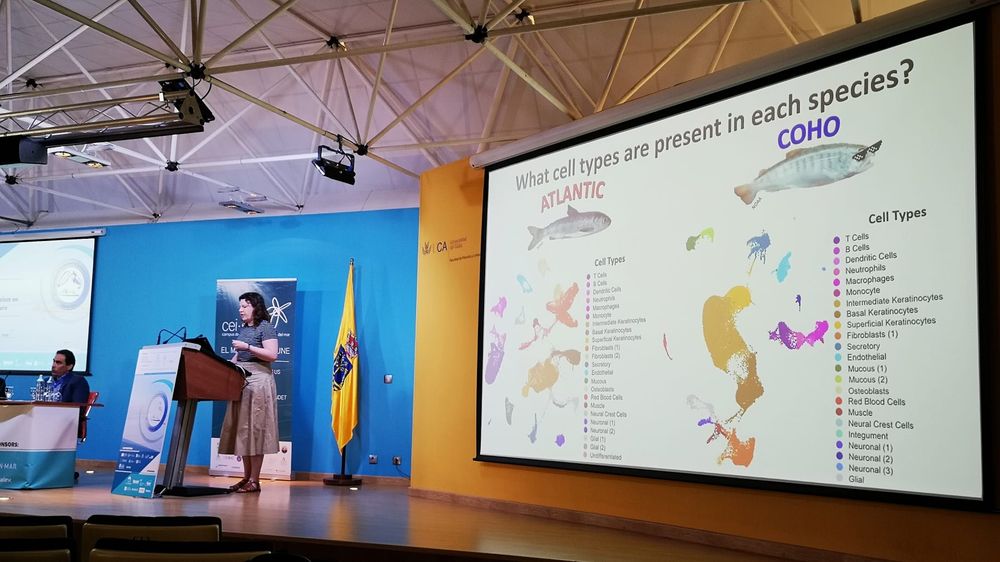
Sarah is leaving the Roslin and starting her group in @exeter.ac.uk, she will be missed !
🧪🧬🖥️

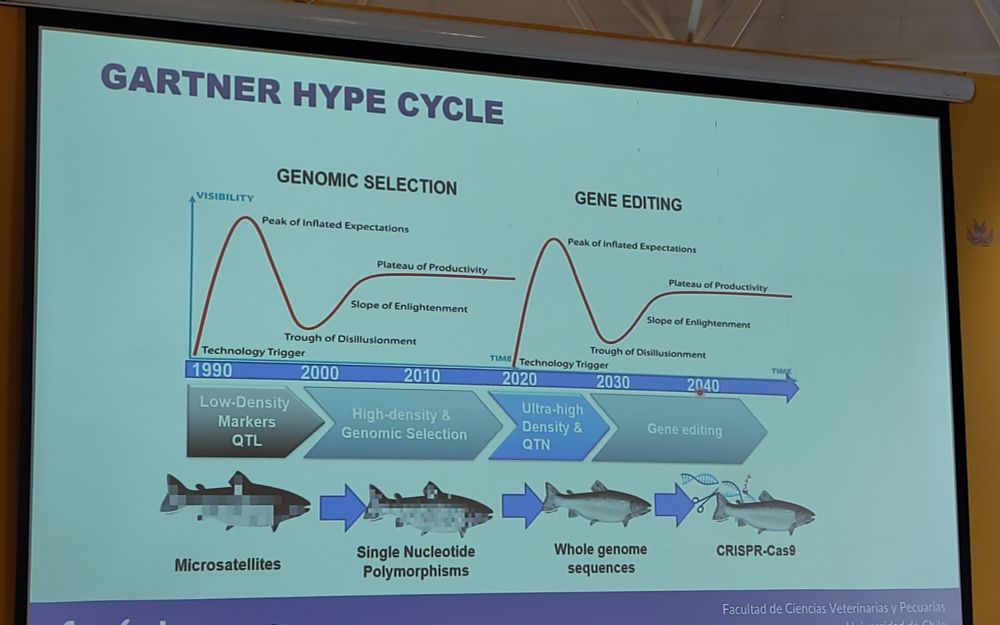
🧪🧬🖥️
There are a few posters as well to look at during the breaks and poster session 👀
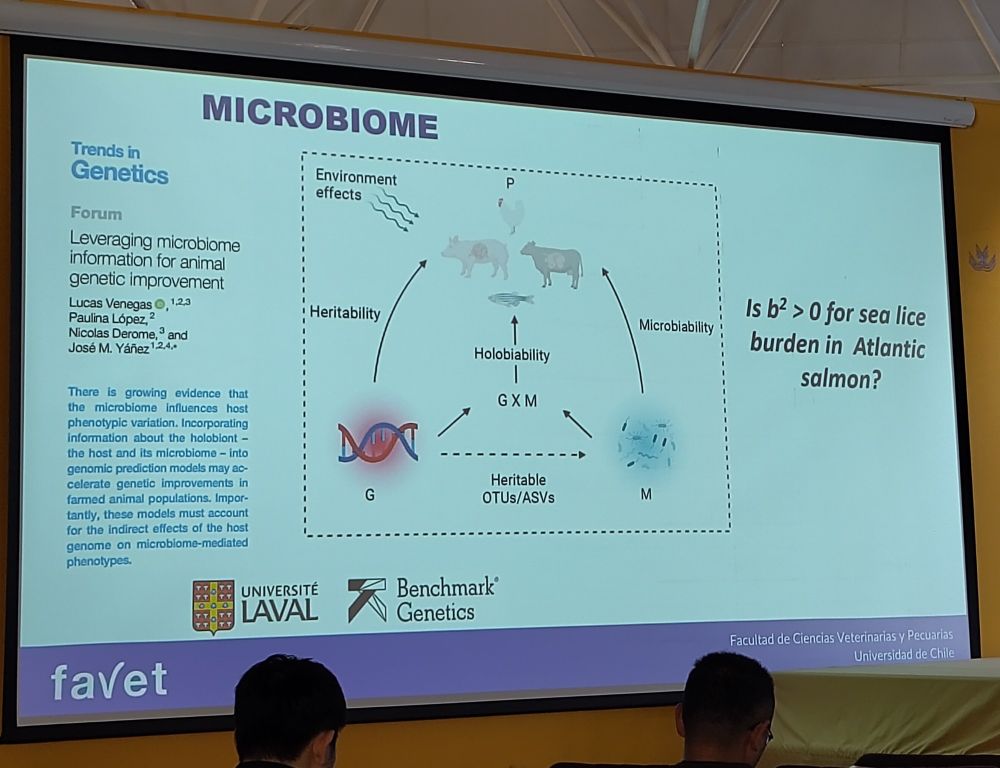
There are a few posters as well to look at during the breaks and poster session 👀
🧬🖥️
🧬🖥️
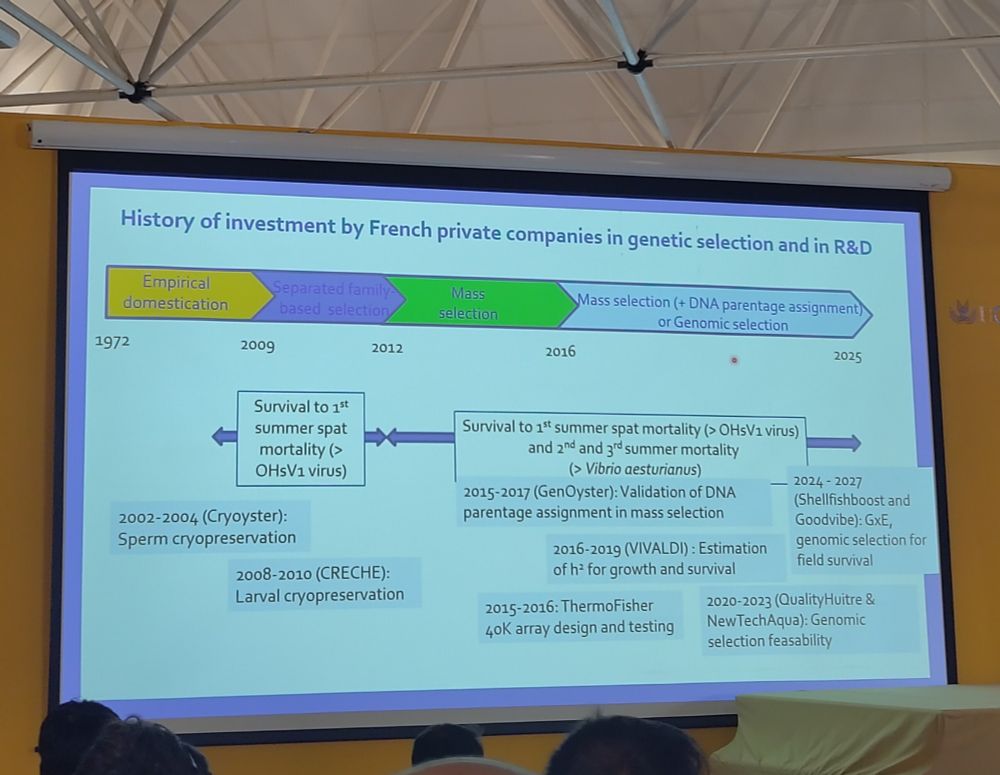

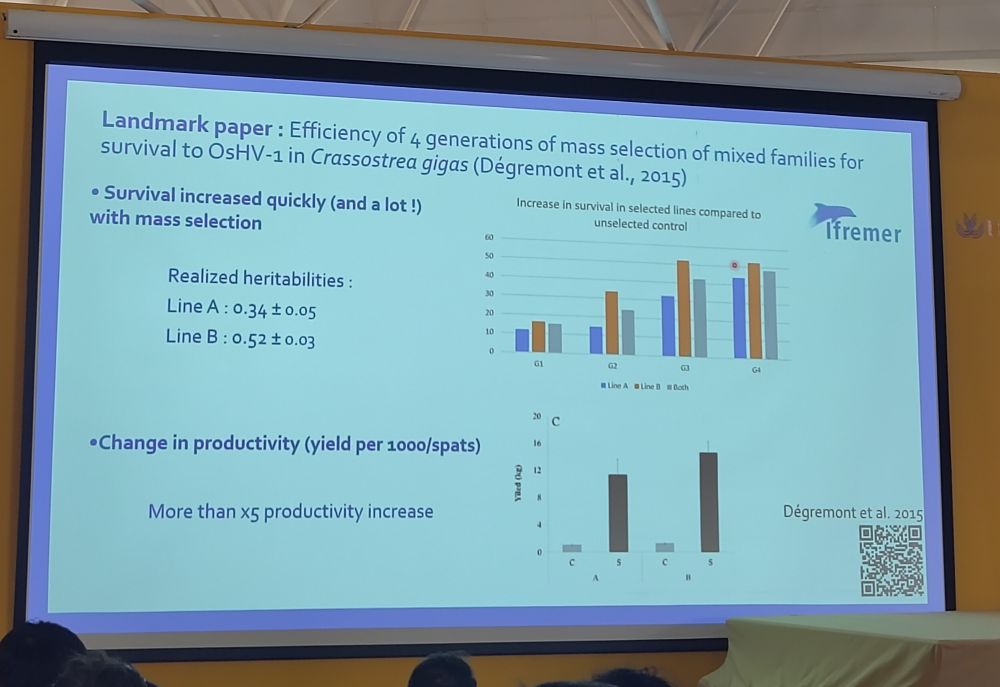
A super clever approach! I can't wait to read the publication! 🧬🐟
A super clever approach! I can't wait to read the publication! 🧬🐟