#GeneRegulation #Scenic+ #NFIB #Transplantation
(6/n)
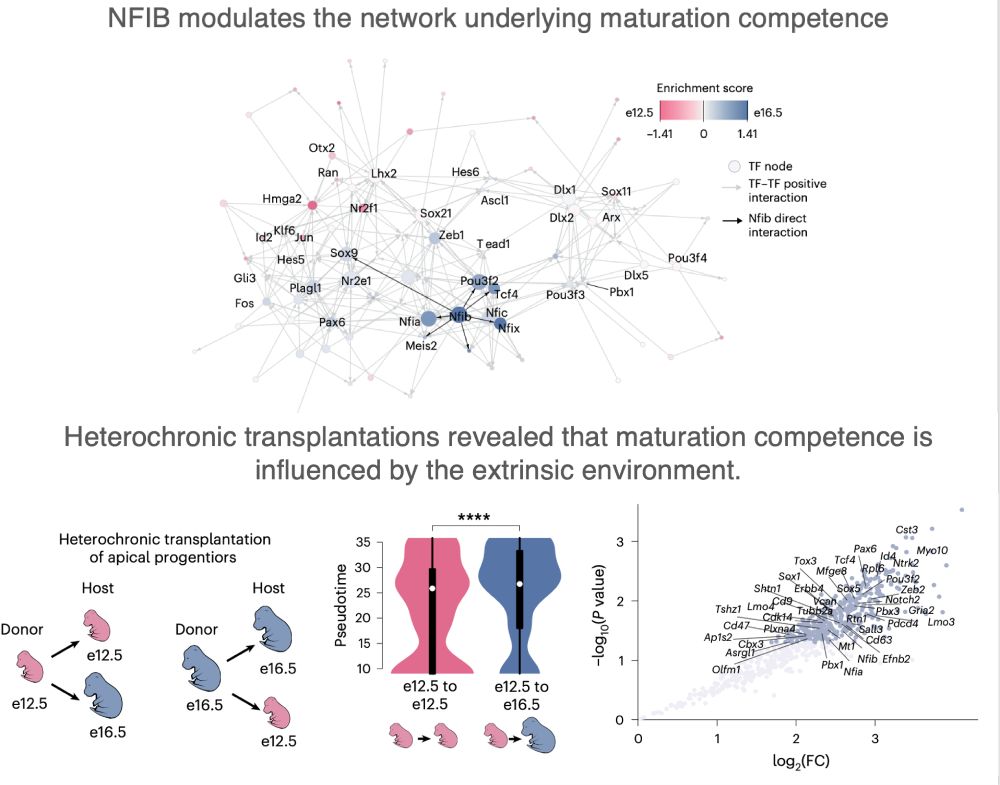
#GeneRegulation #Scenic+ #NFIB #Transplantation
(6/n)
Footprinting showed increased NFI binding, especially NFIB, in late-born cohorts. These neurons activate maturation modules faster — possibly guided by chromatin remodeling during neurogenesis.
#TFfootprints #TranscriptionFactors #scATACseq
(5/n)
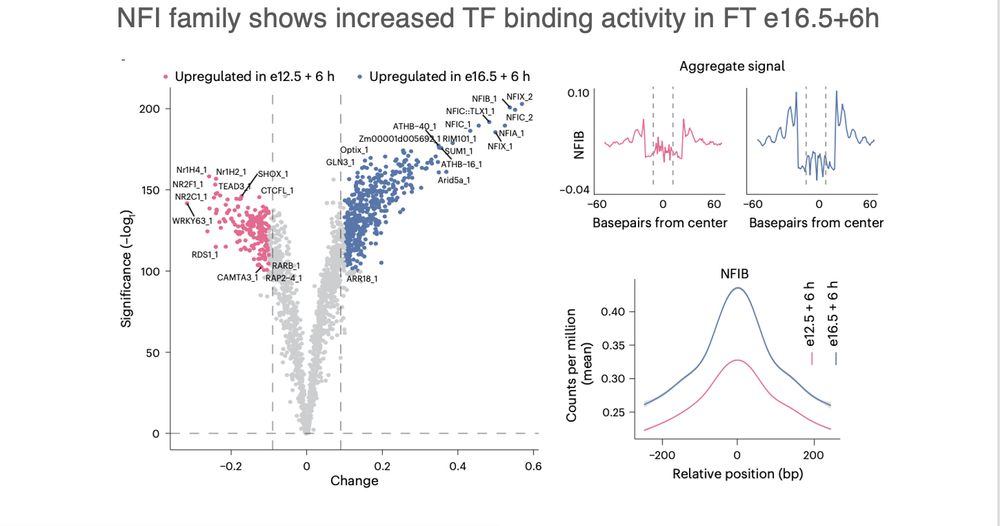
Footprinting showed increased NFI binding, especially NFIB, in late-born cohorts. These neurons activate maturation modules faster — possibly guided by chromatin remodeling during neurogenesis.
#TFfootprints #TranscriptionFactors #scATACseq
(5/n)
scATAC-seq revealed stage-specific enhancer activation, with late-born cells opening distinct regulatory elements — especially in distal regions. A dynamic change in the regulatory landscape.
#Chromatin #Epigenetics #scATACseq
(4/n)
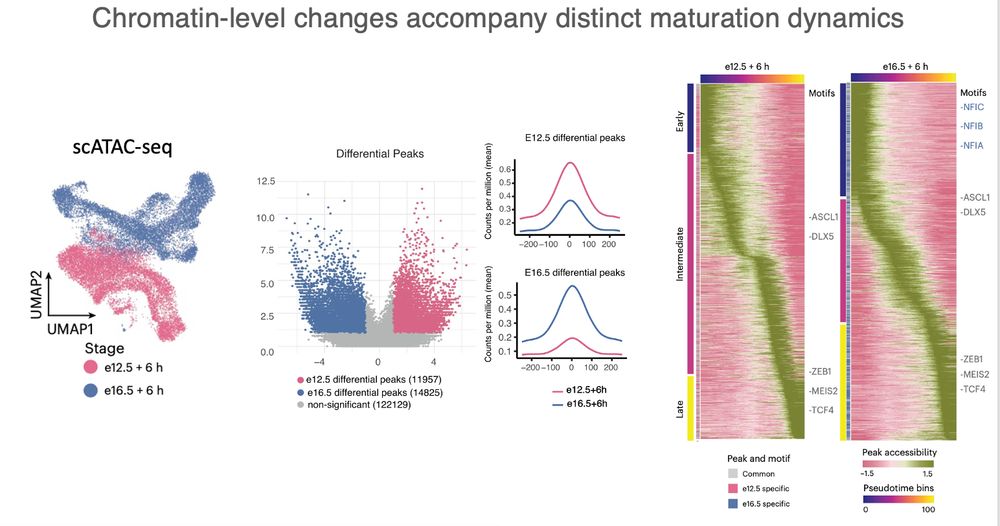
scATAC-seq revealed stage-specific enhancer activation, with late-born cells opening distinct regulatory elements — especially in distal regions. A dynamic change in the regulatory landscape.
#Chromatin #Epigenetics #scATACseq
(4/n)
(3/n)
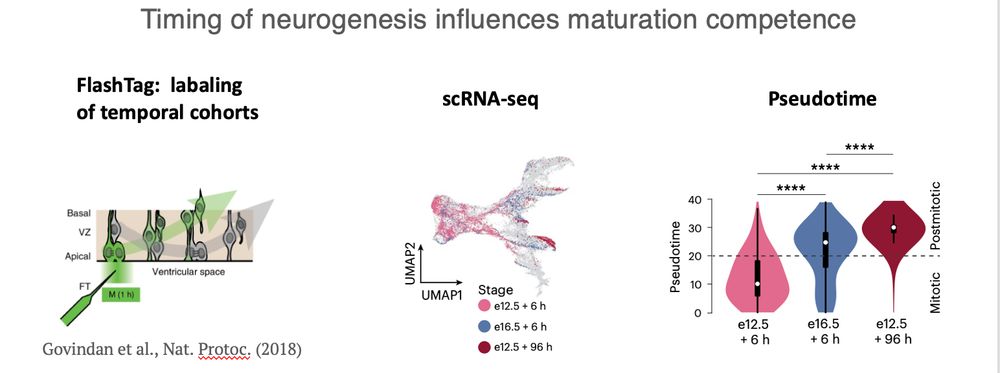
(3/n)
But it turns out that in inhibitory lineages, progenitors retain more stable differentiation competence. Instead, temporal progression alters maturation dynamics.
#PatchClamp
(2/n)
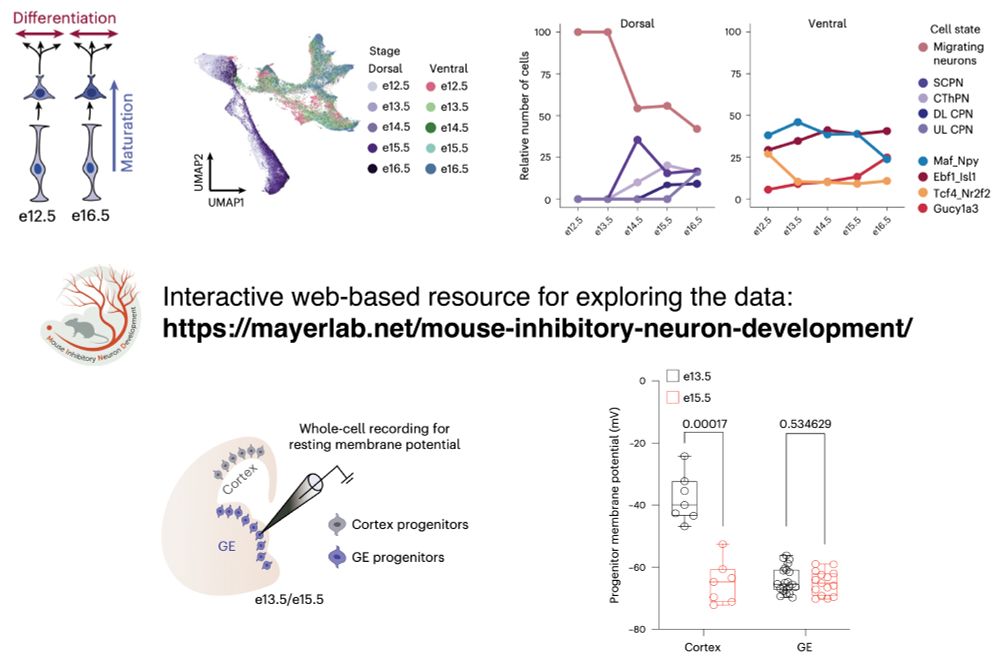
But it turns out that in inhibitory lineages, progenitors retain more stable differentiation competence. Instead, temporal progression alters maturation dynamics.
#PatchClamp
(2/n)

