
http://www.dickinsonlab.uchicago.edu/
bsky.app/profile/chem...

bsky.app/profile/chem...

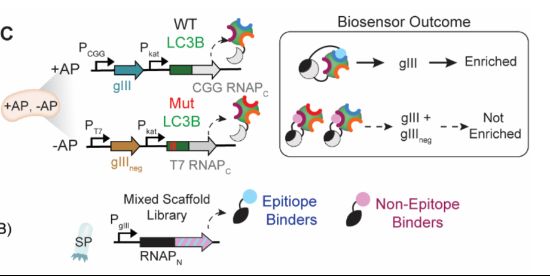
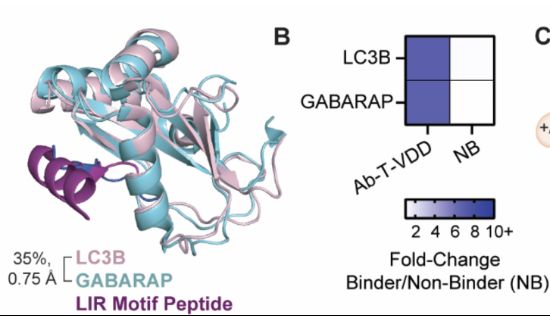



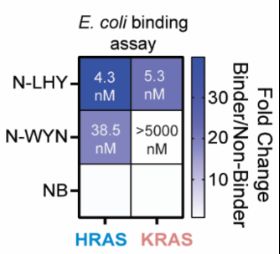


www.nature.com/articles/s41...
3/n

www.nature.com/articles/s41...
3/n



