Yet, PLM or hybrid approaches (even with data-leakage inflating performance) provide little benefit over the best alignment-based model (EVE).

Yet, PLM or hybrid approaches (even with data-leakage inflating performance) provide little benefit over the best alignment-based model (EVE).
Trained on 1M+ new species, it designs novel antibiotics with 97% success. Biodiversity at scale = models that generalize and design.
bit.ly/3NpMR4B

Trained on 1M+ new species, it designs novel antibiotics with 97% success. Biodiversity at scale = models that generalize and design.
bit.ly/3NpMR4B
Alignoth generates self-contained interactive HTML read alignment plots from BAM files – Rust-based, portable, and ideal for headless workflows.
📄 doi.org/10.1093/bioi...
#bioinformatics #genomics #rust @johanneskoester.bsky.social

Alignoth generates self-contained interactive HTML read alignment plots from BAM files – Rust-based, portable, and ideal for headless workflows.
📄 doi.org/10.1093/bioi...
#bioinformatics #genomics #rust @johanneskoester.bsky.social
Today in @science.org, we introduce Conformational Biasing (CB), a simple and scalable computational method that uses contrastive scoring by inverse folding models to identify conformation-biasing mutations.

Today in @science.org, we introduce Conformational Biasing (CB), a simple and scalable computational method that uses contrastive scoring by inverse folding models to identify conformation-biasing mutations.
arxiv.org/abs/2506.12986

Very worryingly we find that TWAS gets the sign wrong around 1/3 of the time (compared to 50% for pure guessing). You can read more about our analysis here, and what we think is going on 👇
In a new pre-print, we find that TWAS gets the sign wrong around 20-30% of the time!
doi.org/10.64898/202...
1/n

Very worryingly we find that TWAS gets the sign wrong around 1/3 of the time (compared to 50% for pure guessing). You can read more about our analysis here, and what we think is going on 👇

Wrapping htslib is my hello-world but doing it in rust to allow javascript expressions handled by V8 was more substantial project.
The tools and LLMs are now getting quite good:
brentp.github.io/latest/blog/...
let me know what you think
Wrapping htslib is my hello-world but doing it in rust to allow javascript expressions handled by V8 was more substantial project.
The tools and LLMs are now getting quite good:
brentp.github.io/latest/blog/...
let me know what you think
andrewcarroll.github.io/2025/12/23/t...
andrewcarroll.github.io/2025/12/23/t...
🧵
🧵
@francescazfl.bsky.social @jsunn-y.bsky.social @francesarnold.bsky.social @arianemora.bsky.social
Paper: doi.org/10.1093/nar/...
DB: enzengdb.org
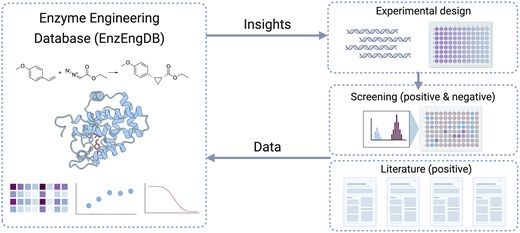
@francescazfl.bsky.social @jsunn-y.bsky.social @francesarnold.bsky.social @arianemora.bsky.social
Paper: doi.org/10.1093/nar/...
DB: enzengdb.org
We've spent years building a system where verification is cheap -- generationally so.
This changes the logic of discovery in ways that are easy to underestimate. I tried to write a little about what that means: www.jurabio.com/blog/onebill...

We've spent years building a system where verification is cheap -- generationally so.
This changes the logic of discovery in ways that are easy to underestimate. I tried to write a little about what that means: www.jurabio.com/blog/onebill...

matsen.group/general/2025...

matsen.group/general/2025...
In an era of AI-designed proteins, the next leap will be controlling when, where, and how much of these proteins are expressed in living cells.
www.biorxiv.org/content/10.1...

In an era of AI-designed proteins, the next leap will be controlling when, where, and how much of these proteins are expressed in living cells.
www.biorxiv.org/content/10.1...


1/n

No RNA-seq.
No protein homology.
No repeats, hints, or curated evidence.
Raw genome → accurate gene models.
Deep learning + HMM, published in @natmethods.nature.com
www.nature.com/articles/s41...

No RNA-seq.
No protein homology.
No repeats, hints, or curated evidence.
Raw genome → accurate gene models.
Deep learning + HMM, published in @natmethods.nature.com
www.nature.com/articles/s41...

We built a proteome-wide model that combines cross-species and human population variation to rank missense variants by disease severity and help diagnose rare genetic disorders.
rdcu.be/eRu7K

We built a proteome-wide model that combines cross-species and human population variation to rank missense variants by disease severity and help diagnose rare genetic disorders.
rdcu.be/eRu7K
pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
matsen.group/agentic.html details:
• How agents work (& why it matters)
• Git Flow with agents
• Using agents for science
• The human-agent interface
Questions? What has your experience been?

matsen.group/agentic.html details:
• How agents work (& why it matters)
• Git Flow with agents
• Using agents for science
• The human-agent interface
Questions? What has your experience been?

