
7) GAME enables continual benchmarking; we anticipate a yearly “state of the field” to be easily produced. Here's an example for gene expression prediction using existing GAME modules.
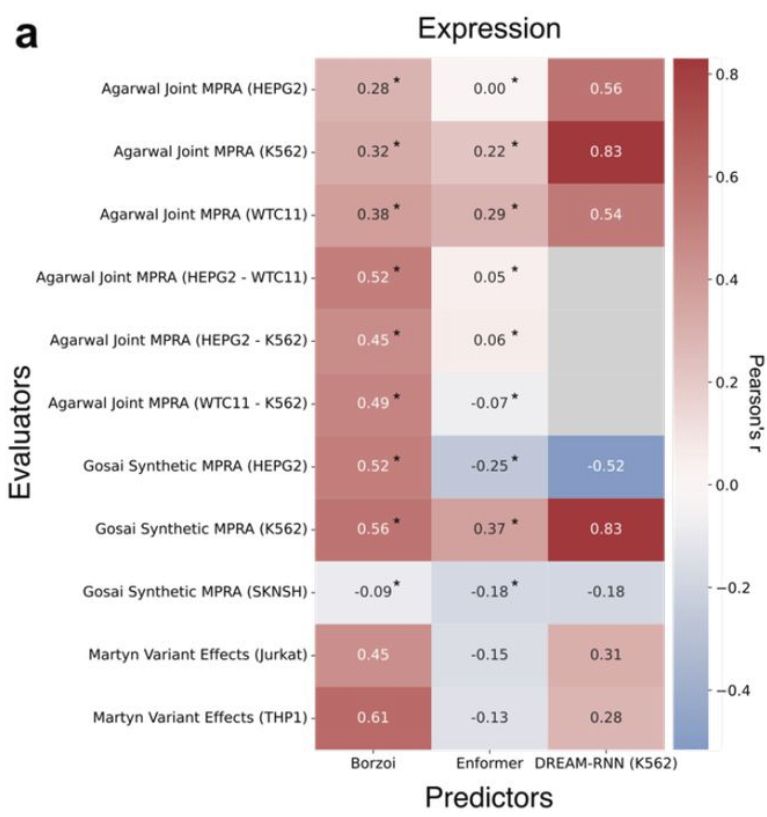
7) GAME enables continual benchmarking; we anticipate a yearly “state of the field” to be easily produced. Here's an example for gene expression prediction using existing GAME modules.
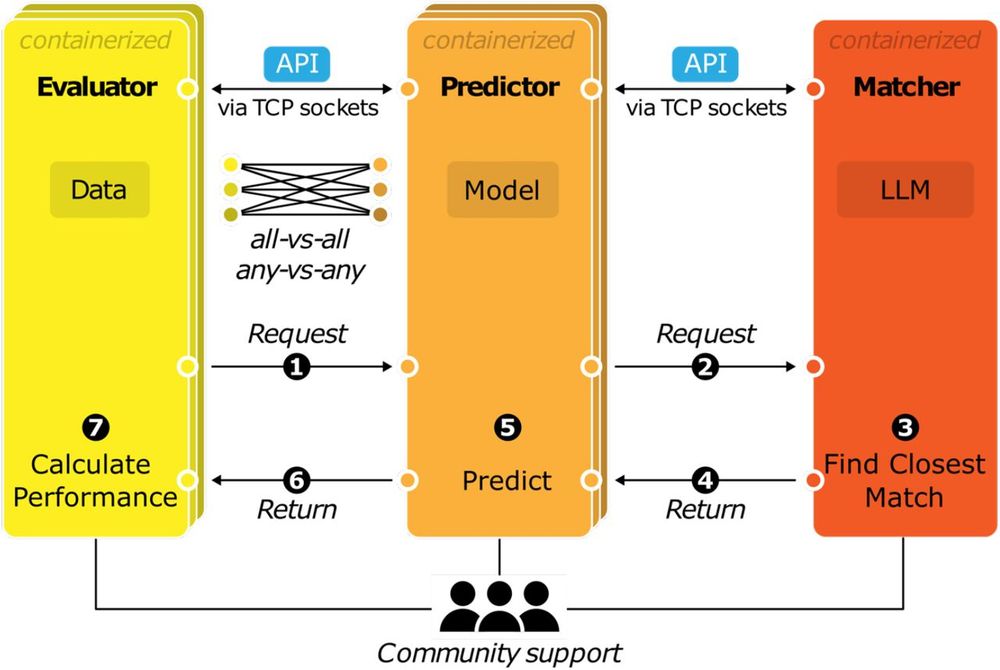
1) Via the API, all models are inherently compatible with all benchmarks
2) GAME will work across platforms – just get Apptainer installed, and away you go 💻↔️🖥️
1) Via the API, all models are inherently compatible with all benchmarks
2) GAME will work across platforms – just get Apptainer installed, and away you go 💻↔️🖥️


de-boer-lab.github.io/CAGT_meeting/

de-boer-lab.github.io/CAGT_meeting/
de-boer-lab.github.io/CAGT_meeting/

de-boer-lab.github.io/CAGT_meeting/
ubc.wd10.myworkdayjobs.com/en-US/ubcfac...

ubc.wd10.myworkdayjobs.com/en-US/ubcfac...


