
https://venturalaboratory.com/
https://www.mskcc.org/research/ski/labs/andrea-ventura
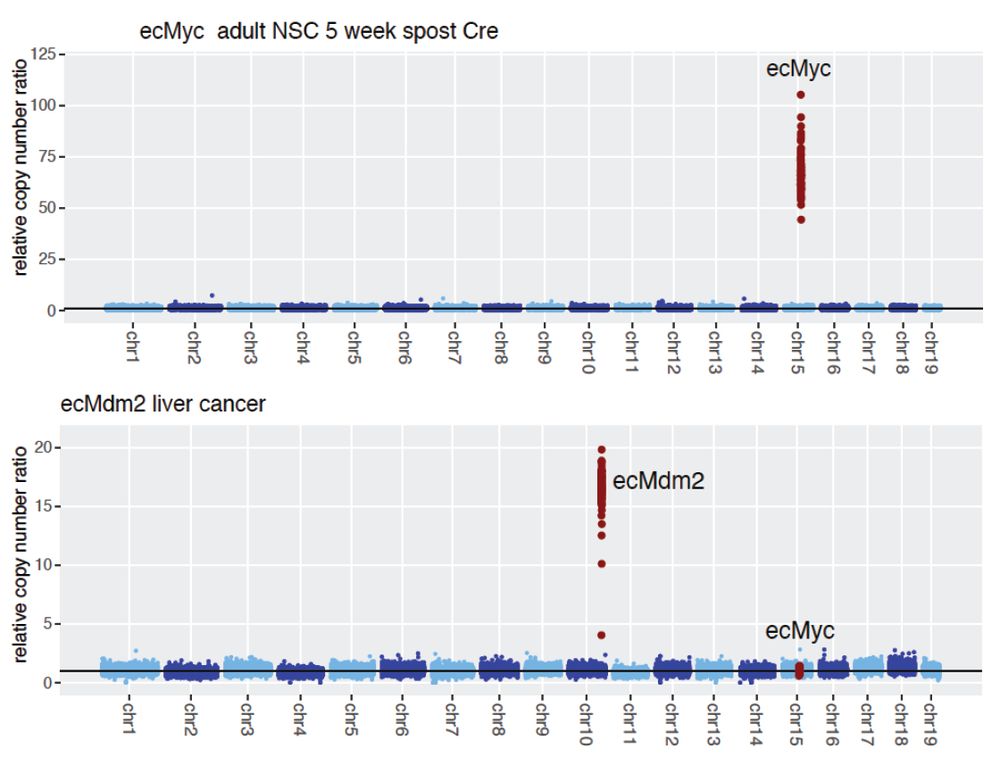
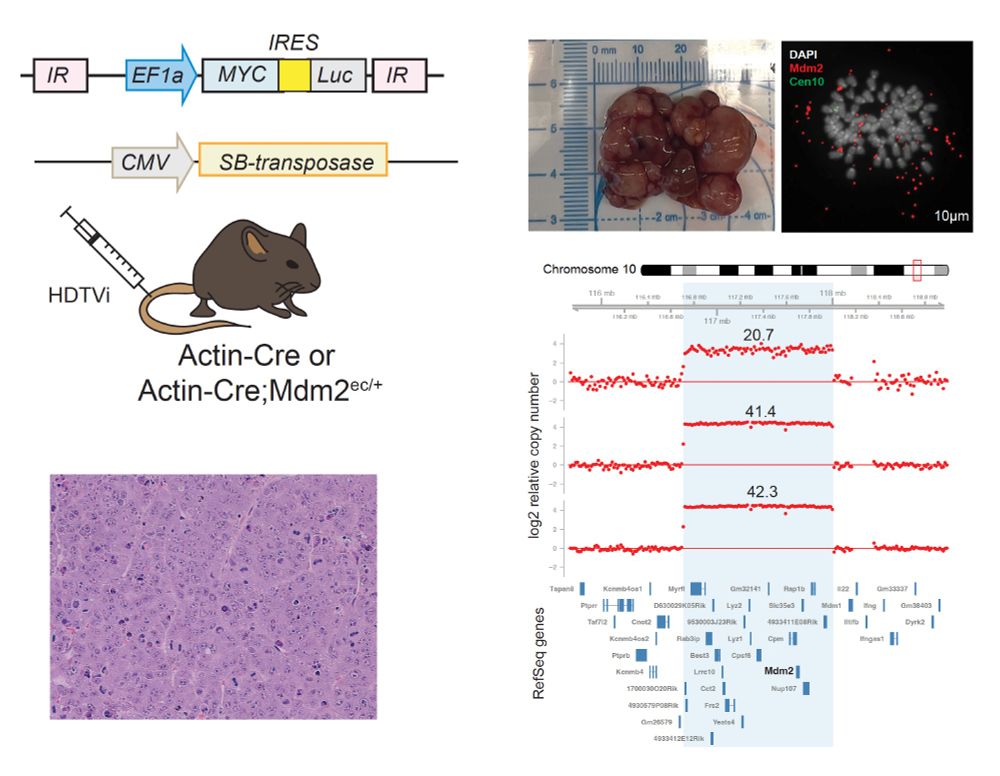






To test this strategy, we engineered the formation of a 1.5Mbp ecDNA encompassing the MDM2 locus (as well as a bunch of other genes) in HCT116 cells, which are otherwise diploid and chromosomally stable.

To test this strategy, we engineered the formation of a 1.5Mbp ecDNA encompassing the MDM2 locus (as well as a bunch of other genes) in HCT116 cells, which are otherwise diploid and chromosomally stable.
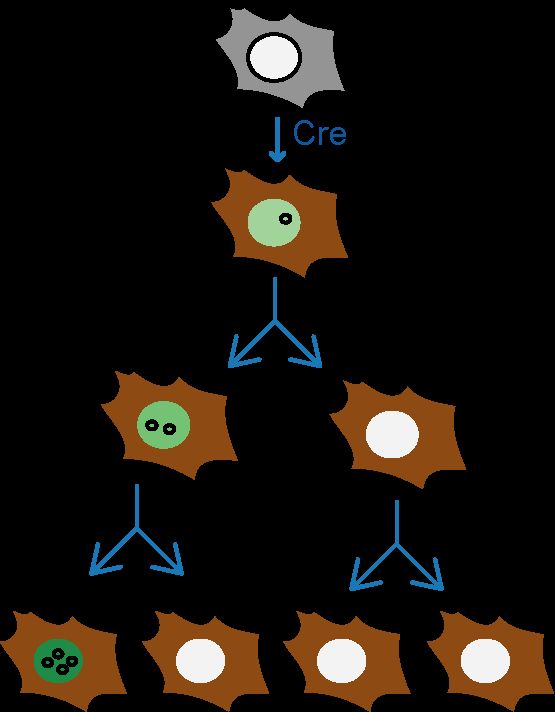
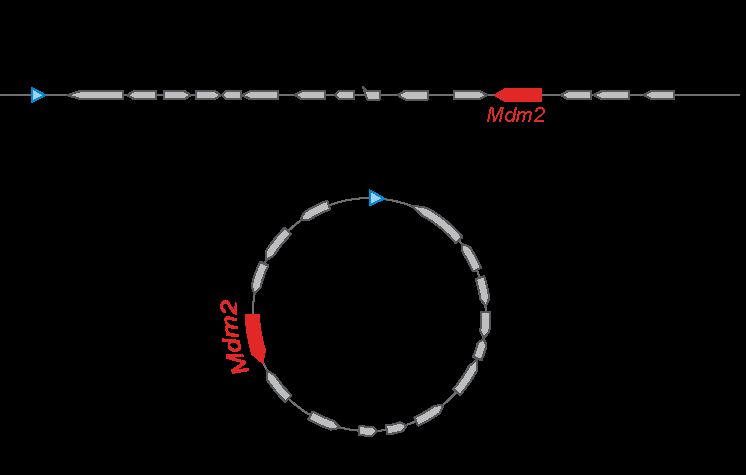
ecDNAs had been known since the 60s (they were called "Double Minutes"). They are large circular DNA fragments found in some of the most aggressive forms of cancers. They amplify oncogenes, boost gene expression, facilitate tumor evolution, and increase resistance to therapy.
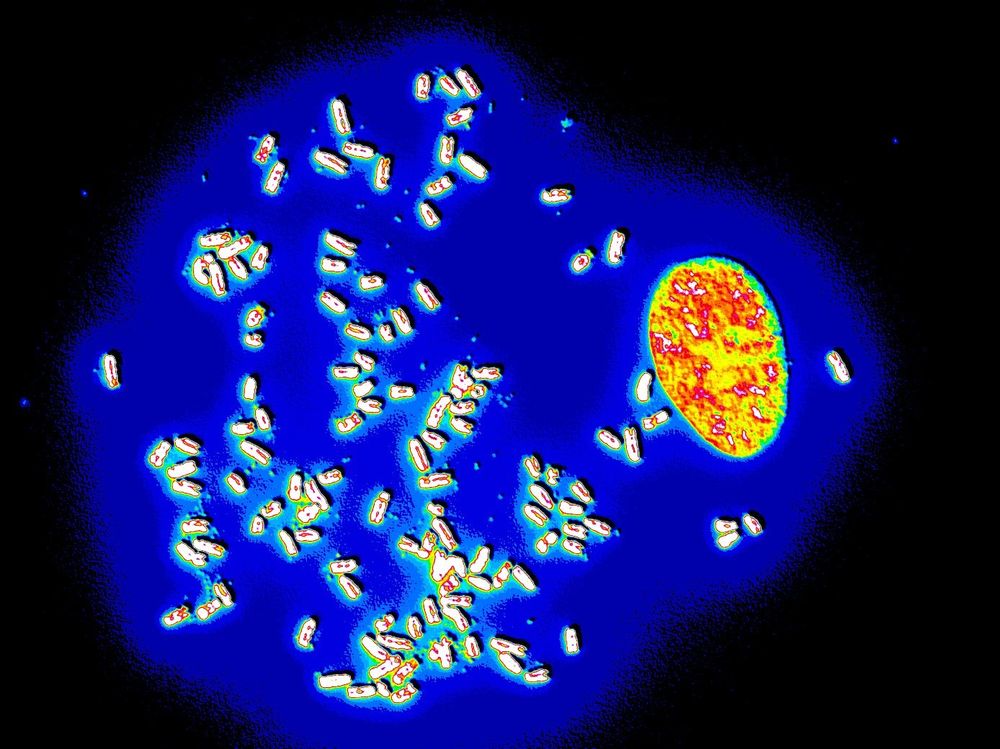
ecDNAs had been known since the 60s (they were called "Double Minutes"). They are large circular DNA fragments found in some of the most aggressive forms of cancers. They amplify oncogenes, boost gene expression, facilitate tumor evolution, and increase resistance to therapy.




