
Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...
Details below
careersearch.stanford.edu/jobs/computa...
Plz RT


Details below
careersearch.stanford.edu/jobs/computa...
Plz RT
www.medrxiv.org/content/10.1...
Saturation genome editing of BRCA1 across cell types accurately resolves cancer risk.
Led by the amazing Phoebe Dace. This one’s packed full of data, so check out the paper. Quick highlights… 🧵 1/n

www.medrxiv.org/content/10.1...
Saturation genome editing of BRCA1 across cell types accurately resolves cancer risk.
Led by the amazing Phoebe Dace. This one’s packed full of data, so check out the paper. Quick highlights… 🧵 1/n
Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/

Led by @sherrynyeo.bsky.social, @erinmayc.bsky.social, and friends, we continue our journey to find viral DNA in our favorite place-- the overlooked and discarded reads in existing data! 1/
medrxiv.org/content/10.1101/2025.06.24.25330216
The main output from my PhD is finally public and we’re SUPER excited about the findings! If you’re interested in what we learnt about IBD with a massive 700+ sample sc-eQTL dataset of the gut, read on!

medrxiv.org/content/10.1101/2025.06.24.25330216
The main output from my PhD is finally public and we’re SUPER excited about the findings! If you’re interested in what we learnt about IBD with a massive 700+ sample sc-eQTL dataset of the gut, read on!

www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
This helps me remember what this is all about.
Ironically, it is about the treatment of pain.

This helps me remember what this is all about.
Ironically, it is about the treatment of pain.
Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...

Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...

Using deep learning & scATAC-seq, we studied context-specific variants in disease & evolution, and introduce FLARE for de novo mutations—w/ application to autism-affected families.
doi.org/10.1101/2025...
Let me know if you have any questions. We're excited to see a wide range of proposals, ranging from AI/ML applications, clinical genetics, women's health, and more.
www.ashg.org/meetings/202...

Let me know if you have any questions. We're excited to see a wide range of proposals, ranging from AI/ML applications, clinical genetics, women's health, and more.
www.ashg.org/meetings/202...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Our new perspective article describing the human and non-human primate developmental GTEx projects is now out in
@nature.com! We outline the scope, vision, opportunities and challenges of these projects here:
www.nature.com/articles/s41...
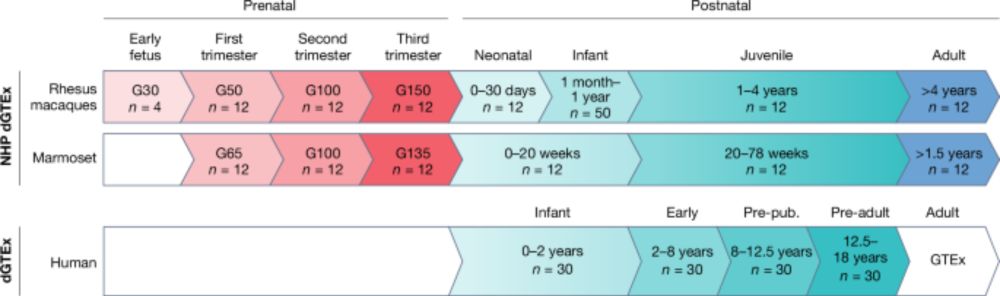
Our new perspective article describing the human and non-human primate developmental GTEx projects is now out in
@nature.com! We outline the scope, vision, opportunities and challenges of these projects here:
www.nature.com/articles/s41...
erictopol.substack.com/p/your-lab-t...

erictopol.substack.com/p/your-lab-t...
Each person has their own tightly regulated setpoints. One healthy person's complete blood count setpoint can be differentiated from 98% of other healthy adults.
www.nature.com/articles/s41...
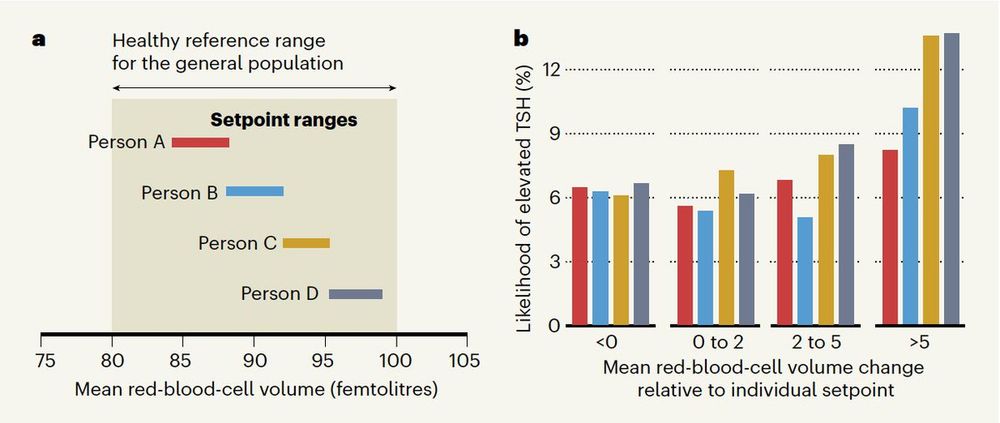
Each person has their own tightly regulated setpoints. One healthy person's complete blood count setpoint can be differentiated from 98% of other healthy adults.
www.nature.com/articles/s41...


Some new answers in our latest preprint, where we explored:
1). Key cell types contributing to CHD genetics
2). Impact of noncoding variants on CHD risk
https://www.medrxiv.org/content/10.1101/2024.11.20.24317557v1
Some new answers in our latest preprint, where we explored:
1). Key cell types contributing to CHD genetics
2). Impact of noncoding variants on CHD risk
https://www.medrxiv.org/content/10.1101/2024.11.20.24317557v1
Link: doi.org/10.1016/j.cmet.2024.06.006
Thread 🧵👇1/9

Link: doi.org/10.1016/j.cmet.2024.06.006
Thread 🧵👇1/9


