



Maps condensates w/ microfluidics & lysate. Probes stability in stress granules, nucleoli, & FUS mutants.


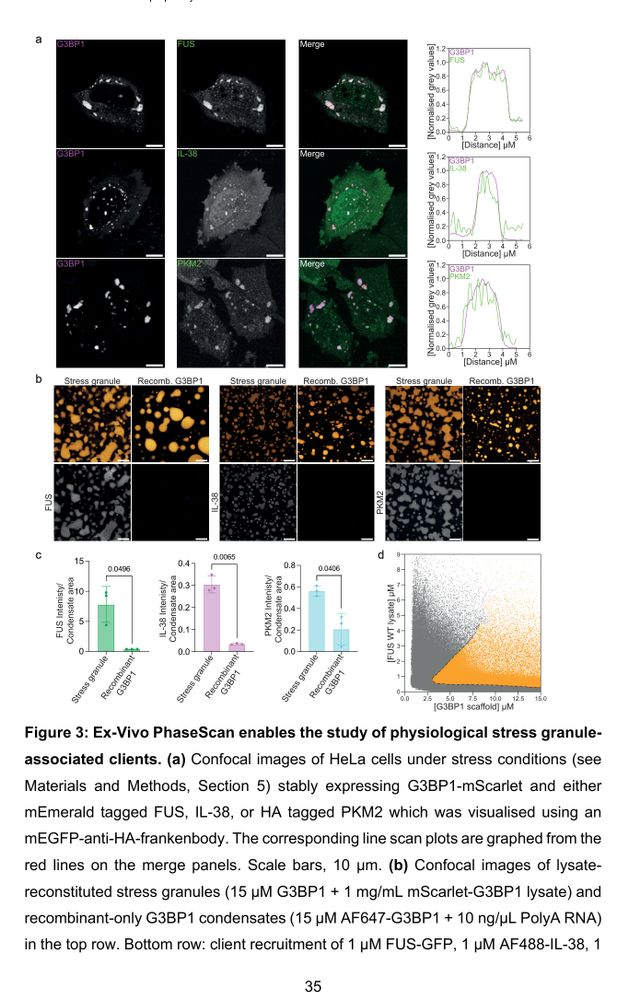

Maps condensates w/ microfluidics & lysate. Probes stability in stress granules, nucleoli, & FUS mutants.
*De novo* protein design potential via LLMs; promising for engineering.



*De novo* protein design potential via LLMs; promising for engineering.
Agentic LLM framework aids mechanistic enzyme design by integrating reasoning, structural analysis, and property prediction to generate testable hypotheses.




Agentic LLM framework aids mechanistic enzyme design by integrating reasoning, structural analysis, and property prediction to generate testable hypotheses.
Generates protein backbones by denoising torsion angles, ensuring ideal local geometry. Refinement optimizes compactness while maintaining bond constraints.




Generates protein backbones by denoising torsion angles, ensuring ideal local geometry. Refinement optimizes compactness while maintaining bond constraints.
GFlowNets open box: learns chem rules, finds key atomic env, prop, motifs in mol design.




GFlowNets open box: learns chem rules, finds key atomic env, prop, motifs in mol design.
Infers cell lineage via SNVs & CNAs jointly for accuracy.




Infers cell lineage via SNVs & CNAs jointly for accuracy.
Inferring cell environments by modeling cell interactions as agents learned via reinforcement learning. Enables in-silico manipulation and prediction.




Inferring cell environments by modeling cell interactions as agents learned via reinforcement learning. Enables in-silico manipulation and prediction.
Deconvolves sparse RNA-seq using NB models, revealing cancer insights.




Deconvolves sparse RNA-seq using NB models, revealing cancer insights.
miRNA-mRNA interactions via dual-transformer; scalable sliding-window attention on sequence pairs.




miRNA-mRNA interactions via dual-transformer; scalable sliding-window attention on sequence pairs.
RL controls bacteria via adaptive treatments based on population data & nutrients.




RL controls bacteria via adaptive treatments based on population data & nutrients.
LumiFAST luciferase: Tunable BRET bioluminescence color w/ fluorogens for deep tissue imaging/biosensing.




LumiFAST luciferase: Tunable BRET bioluminescence color w/ fluorogens for deep tissue imaging/biosensing.




Disentangled learning: infer counterfactuals & estimate treatment effects from single-cell data.




Disentangled learning: infer counterfactuals & estimate treatment effects from single-cell data.
Designs biomolecular binders by unifying structure prediction & design. Enables flexible control & has been experimentally validated across targets.




Designs biomolecular binders by unifying structure prediction & design. Enables flexible control & has been experimentally validated across targets.
CNN encodes motif profiles for cis-regulatory syntax, TF coop., & pluripotency insights.




CNN encodes motif profiles for cis-regulatory syntax, TF coop., & pluripotency insights.
Delayed splicing is a key mechanism for timing inflammatory gene expression. Suboptimal splice sites and regulatory sequences fine-tune transcript maturation.


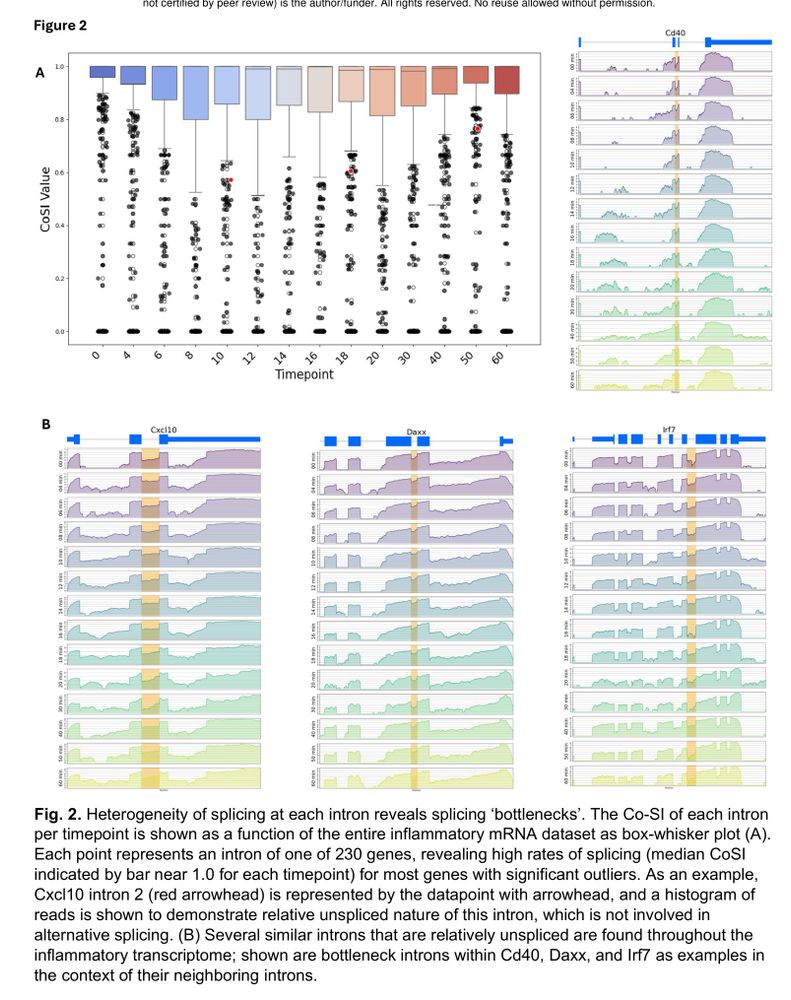

Delayed splicing is a key mechanism for timing inflammatory gene expression. Suboptimal splice sites and regulatory sequences fine-tune transcript maturation.
Cryo-ET: finds protein domains by fitting AlphaFold2 models with Bayesian inference.




Cryo-ET: finds protein domains by fitting AlphaFold2 models with Bayesian inference.
Integrates smooth hydrophobicity/charge with structure for function-oriented protein design, improving sequence recovery and variant generation.




Integrates smooth hydrophobicity/charge with structure for function-oriented protein design, improving sequence recovery and variant generation.
LongPhase-S: Haplotype reconst., infers tumor purity, recalibrates variants for somatic calls in long reads.




LongPhase-S: Haplotype reconst., infers tumor purity, recalibrates variants for somatic calls in long reads.




Greek AI life science research: methods/algorithms, less disease/clinics. Barriers noted.




Greek AI life science research: methods/algorithms, less disease/clinics. Barriers noted.
Finds cell-type genes via axioms, showing specificity & behavior.




Finds cell-type genes via axioms, showing specificity & behavior.
Analyzes differential methylation in WGBS data. It models genomic heterogeneity using context-specific dispersion shrinkage, improving detection accuracy.




Analyzes differential methylation in WGBS data. It models genomic heterogeneity using context-specific dispersion shrinkage, improving detection accuracy.
Detects circRNAs w/ complex repeat patterns, incl. partial rev. comp.




Detects circRNAs w/ complex repeat patterns, incl. partial rev. comp.

