Some bio and coding stuff
Please spread the word; I think many people just outside my own circle could benefit from this :)
cc @rickbitloo.bsky.social
github.com/RagnarGrootK...
Please spread the word; I think many people just outside my own circle could benefit from this :)
cc @rickbitloo.bsky.social
github.com/RagnarGrootK...
Grep mode shows all matches, grouped per record, and is meant for human consumption.
Filter mode prints full matching (or non-matching) records to stdout or output files.

Grep mode shows all matches, grouped per record, and is meant for human consumption.
Filter mode prints full matching (or non-matching) records to stdout or output files.
It's the first tool that builds on Sassy, the approximate-DNA-searching tool that @rickbitloo.bsky.social and myself developed earlier this year, specifically with this application in mind.
www.biorxiv.org/content/10.1...
Want to get started? github.com/rickbeeloo/b...
It's the first tool that builds on Sassy, the approximate-DNA-searching tool that @rickbitloo.bsky.social and myself developed earlier this year, specifically with this application in mind.
www.biorxiv.org/content/10.1...
Want to get started? github.com/rickbeeloo/b...
www.biorxiv.org/content/10.1...
Want to get started? github.com/rickbeeloo/b...
Ever need to search for approximate matches of short DNA strings?
Sassy is the tool to use!
Available now wherever you get your code
With @rickbitloo.bsky.social
curiouscoding.nl/papers/sassy...
github.com/ragnarGrootK...
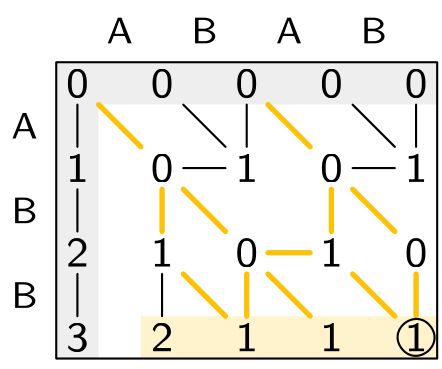
Ever need to search for approximate matches of short DNA strings?
Sassy is the tool to use!
Available now wherever you get your code
With @rickbitloo.bsky.social
curiouscoding.nl/papers/sassy...
github.com/ragnarGrootK...

