Nitesh Khandelwal
@nkhandelwal.bsky.social
66 followers
210 following
11 posts
Research Assistant Professor at OHSU.
Structure biology| Drug resistance| Transporter| Cryo-EM| Membrane proteins|Microbiology|
Past: #UCSF, Research Specialist II | Bio5 postdoctoral fellow @uaarizona | #ICGEB #JNU #MSU
Posts
Media
Videos
Starter Packs
Reposted by Nitesh Khandelwal
Reposted by Nitesh Khandelwal
Shanti Gangwar
@shantigangwar.bsky.social
· Aug 22

Activation of kainate receptor GluK2–Neto2 complex - Nature Structural & Molecular Biology
Gangwar et al. present cryo-electron microscopy structures of GluK2 kainate receptors with auxiliary subunit Neto2 and reveal the basis of receptor activation.
www.nature.com
cryoEM papers
@cryoempapers.bsky.social
· Jul 19
Reposted by Nitesh Khandelwal
Meghna Gupta
@meghnag27.bsky.social
· May 27
Reposted by Nitesh Khandelwal
Meghna Gupta
@meghnag27.bsky.social
· May 27
Reposted by Nitesh Khandelwal
Cornelius Gati
@gati.bsky.social
· Apr 23
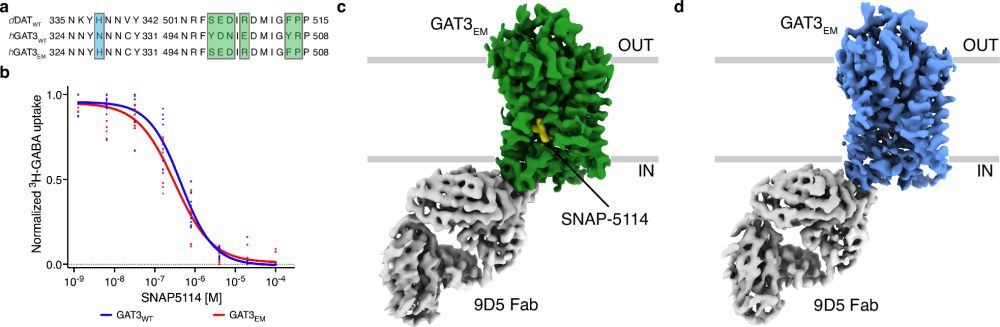
Molecular basis of human GABA transporter 3 inhibition
Nature Communications - GABA transporters maintain GABA homeostasis and regulate synaptic signaling. Here, the authors determined the cryoEM structures of GABA transporter 3 (GAT3) in its apo form...
urldefense.com
Reposted by Nitesh Khandelwal
Reposted by Nitesh Khandelwal
Reposted by Nitesh Khandelwal
Reposted by Nitesh Khandelwal
Reposted by Nitesh Khandelwal
Reposted by Nitesh Khandelwal
Reposted by Nitesh Khandelwal
Reichow Lab
@reichowlab.bsky.social
· Feb 21