https://mecfsscience.org/
"We hypothesize that by conducting a fecal microbiota transplantation (FMT) with stool from ME/CFS cases into mice that are engrafted with a functional human immune system, we can develop an animal model that will replicate ME/CFS-associated pathology."

"We hypothesize that by conducting a fecal microbiota transplantation (FMT) with stool from ME/CFS cases into mice that are engrafted with a functional human immune system, we can develop an animal model that will replicate ME/CFS-associated pathology."
Goes through many examples such as Herpes simplex, Hepatitis B and C, Ebola, SSPE in measles, Varicella-zoster, and EBV.

Goes through many examples such as Herpes simplex, Hepatitis B and C, Ebola, SSPE in measles, Varicella-zoster, and EBV.
The infrastructure first built for HIV, then used for Long Covid in the LIINC study, will now also help with studying pre-pandemic ME/CFS
The research will focus on gut biopsies and PET-CT scans.
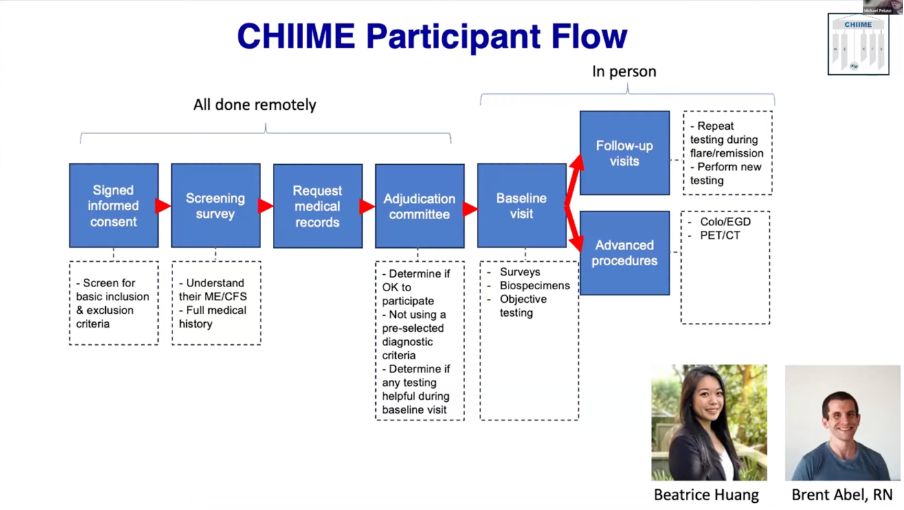
The infrastructure first built for HIV, then used for Long Covid in the LIINC study, will now also help with studying pre-pandemic ME/CFS
The research will focus on gut biopsies and PET-CT scans.
www.medscape.com/viewarticle/...
Lots of good tips to deal with insurance companies.
Government schemes can vary by country but people in most countries can probably pick up useful tips from this also.
#MEcfs #LongCovid #CFS #PwME

www.medscape.com/viewarticle/...
Lots of good tips to deal with insurance companies.
Government schemes can vary by country but people in most countries can probably pick up useful tips from this also.
#MEcfs #LongCovid #CFS #PwME
He suspects there is an inefficiency in producing ATP (energy), but this isn’t only about mitochondria (the engines), but also about the fuel they get...

He suspects there is an inefficiency in producing ATP (energy), but this isn’t only about mitochondria (the engines), but also about the fuel they get...

He suspects there is an inefficiency in producing ATP (energy), but this isn’t only about mitochondria (the engines), but also about the fuel they get...

While we have doubts about their strong focus on viral persistence, their studies on brain scans and tissue samples look very interesting. 🧵

While we have doubts about their strong focus on viral persistence, their studies on brain scans and tissue samples look very interesting. 🧵
They highlight the feeling of being trapped by the illness, profound isolation and longing for the outside world.

They highlight the feeling of being trapped by the illness, profound isolation and longing for the outside world.
Antibodies from controls, serum of Long Covid patients, or papain-digested antibodies did not have the same effect.

Antibodies from controls, serum of Long Covid patients, or papain-digested antibodies did not have the same effect.
The study used data from the National Health Interview Survey (NHIS), however, which has some big limitations...

The study used data from the National Health Interview Survey (NHIS), however, which has some big limitations...
Applicants should submit an Outline Proposal Form before Friday, 9 January 2026.

Applicants should submit an Outline Proposal Form before Friday, 9 January 2026.
Of those infected with SARS-CoV-2 infection, 5% had persistently high Long COVID-related symptom burden. An additional 12% had a high burden that fluctuated but did not improve over time.

Of those infected with SARS-CoV-2 infection, 5% had persistently high Long COVID-related symptom burden. An additional 12% had a high burden that fluctuated but did not improve over time.
The first (Dr. Shady Younis) and last author of the paper (Dr. William H. Robinson) have previously been involved in ME/CFS research.

The first (Dr. Shady Younis) and last author of the paper (Dr. William H. Robinson) have previously been involved in ME/CFS research.
This was presented at the International Conference in Portugal earlier this week.

This was presented at the International Conference in Portugal earlier this week.
Pacing came out on top
#MyalgicEncephalomyelitis #ChronicFatigueSyndrome #MEcfs #CFS #PwME

Pacing came out on top
#MyalgicEncephalomyelitis #ChronicFatigueSyndrome #MEcfs #CFS #PwME
It found altered patterns in ME/CFS patients compared to controls, particularly in proteins involved in the immune system, signal transduction, and muscle contraction.

It found altered patterns in ME/CFS patients compared to controls, particularly in proteins involved in the immune system, signal transduction, and muscle contraction.
Beyond post-viral diseases like ME/CFS and Long COVID — for which effective therapy is urgently needed — viral infections are also implicated in diseases like MS, rheumatoid arthritis,
...
They are calling it "The National Decade Against Post-Infectious Diseases"

Beyond post-viral diseases like ME/CFS and Long COVID — for which effective therapy is urgently needed — viral infections are also implicated in diseases like MS, rheumatoid arthritis,
...
They are calling it "The National Decade Against Post-Infectious Diseases"

They are calling it "The National Decade Against Post-Infectious Diseases"
Unfortunately, it's almost fully based on self-reported data and questionnaires.
There are still almost no studies that looked deeper into MCAS in ME/CFS using proper biological tests.

Unfortunately, it's almost fully based on self-reported data and questionnaires.
There are still almost no studies that looked deeper into MCAS in ME/CFS using proper biological tests.
Too many findings to mention them all, but here are some interesting results...

DecodeME and the Snyder preprint also pointed in that direction (although using other genes and approaches).
In female patients, these were genes involved in neuronal differentiation and development such as ZNF469, BRINP2, and FEZF2.

DecodeME and the Snyder preprint also pointed in that direction (although using other genes and approaches).
Too many findings to mention them all, but here are some interesting results...

Too many findings to mention them all, but here are some interesting results...
It tested:
- computerized cognitive training
- cognitive-behavioral rehabilitation
- transcranial direct current stimulation
but no intervention showed significant improvement compared to a control group that did video puzzles and games.

It tested:
- computerized cognitive training
- cognitive-behavioral rehabilitation
- transcranial direct current stimulation
but no intervention showed significant improvement compared to a control group that did video puzzles and games.

