lindsey guan
@lindseyguan.bsky.social
680 followers
140 following
10 posts
she/her | PhD student in the Keating Lab @MIT | Bio & CS @UCBerkeley
Posts
Media
Videos
Starter Packs
Reposted by lindsey guan
Nature
@nature.com
· 20d

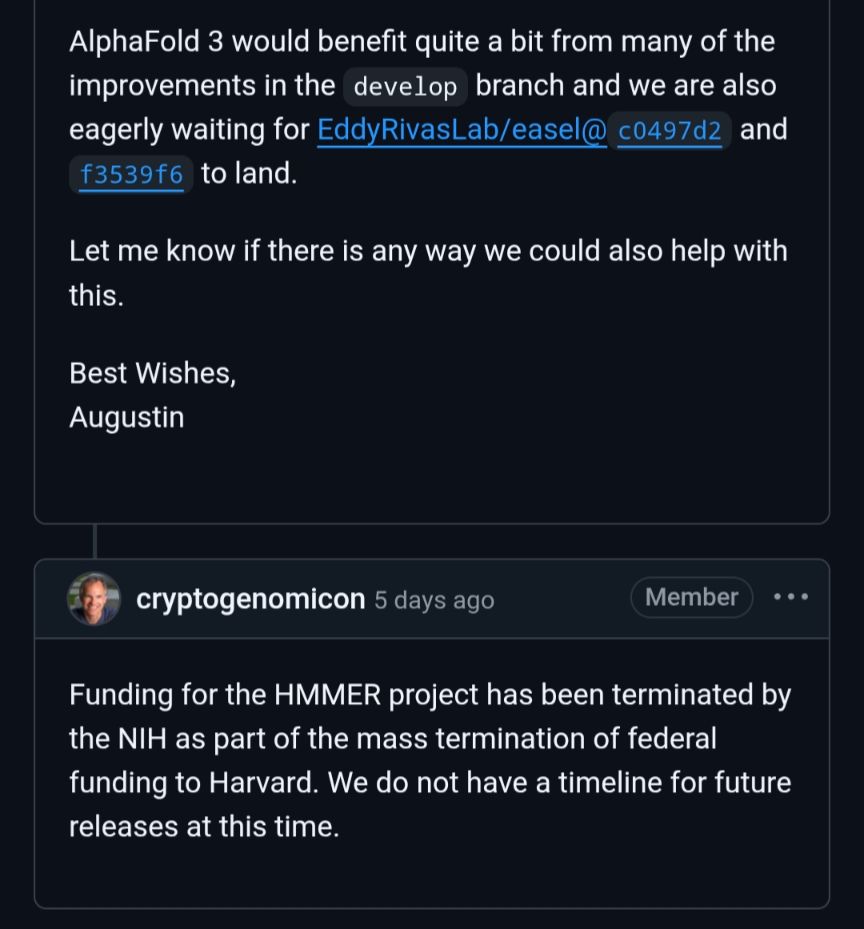
What research might be lost after the NIH’s cuts? Nature trained a bot to find out
We used machine-learning tools in an attempt to recreate the method for cutting funding, and then applied it to past US National Institutes of Health grants to reveal the broad-reaching consequences of such action.
go.nature.com
Reposted by lindsey guan
Reposted by lindsey guan
Yo Akiyama
@yoakiyama.bsky.social
· Aug 5

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
biorxiv.org
Reposted by lindsey guan
Reposted by lindsey guan
Reposted by lindsey guan
Reposted by lindsey guan
Reposted by lindsey guan
Eli Lipsitz
@eli.lipsitz.net
· Feb 12

Game Bub: open-source FPGA retro emulation handheld
I’m excited to announce the project I’ve been working on for the last year and a half: Game Bub, an open-source FPGA based retro emulation handheld, with support for Game Boy, Game Boy Color, and Game...
eli.lipsitz.net
Reposted by lindsey guan
Reposted by lindsey guan
lindsey guan
@lindseyguan.bsky.social
· Nov 27

Jointly Embedding Protein Structures and Sequences through Residue Level Alignment
Residue Level Alignment integrates protein sequence and structure information in a self-supervised model, improving speed and precision in predicting protein binding and structural stability.
journals.aps.org