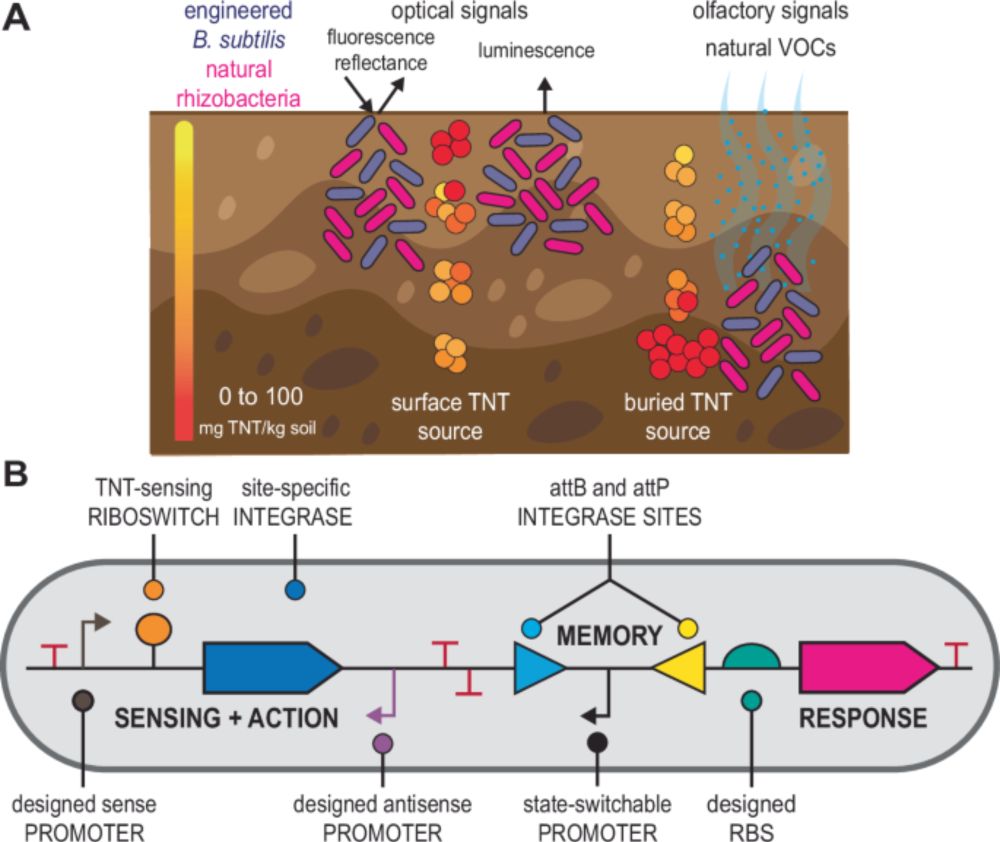
Post-doctoral fellow in the Genome Institute of Singapore.
www.nature.com/articles/s41...

www.nature.com/articles/s41...
lh3.github.io/2025/09/11/a...

lh3.github.io/2025/09/11/a...
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
@unistra.fr @cnrsbiologie.bsky.social
www.nature.com/articles/s41...
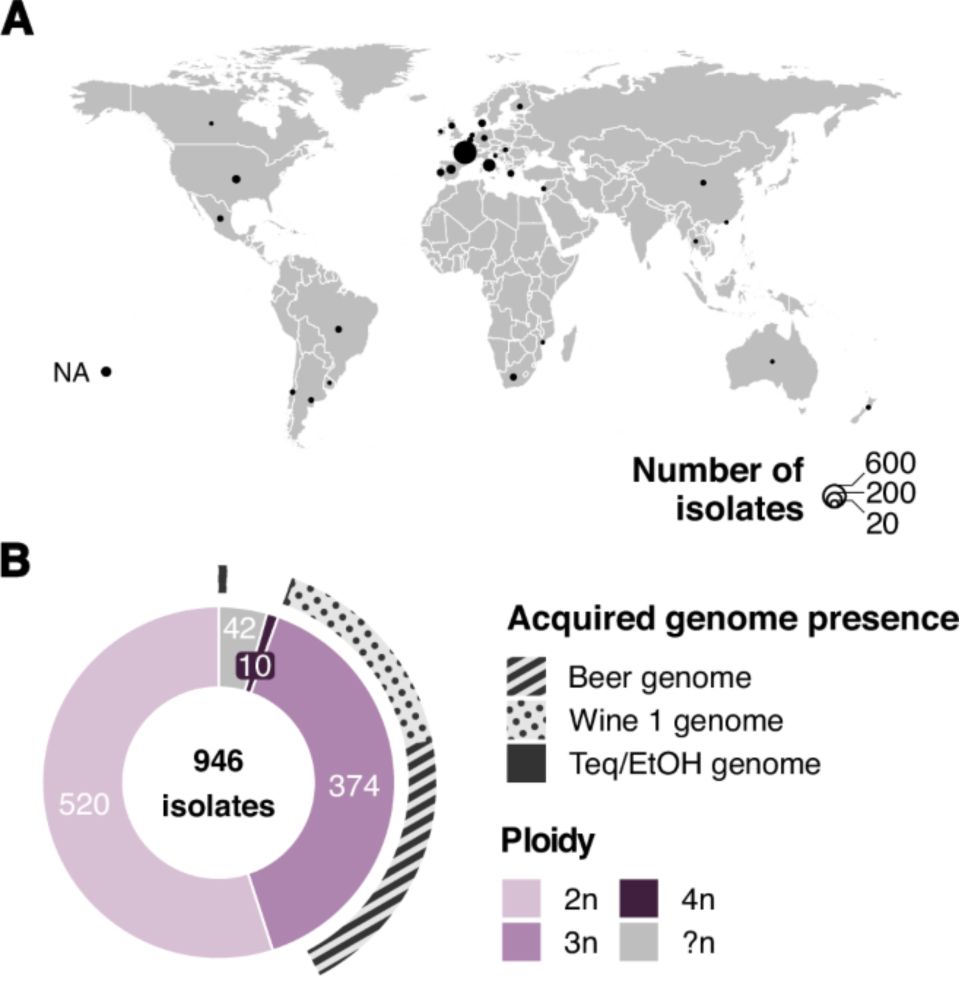
@unistra.fr @cnrsbiologie.bsky.social
www.nature.com/articles/s41...
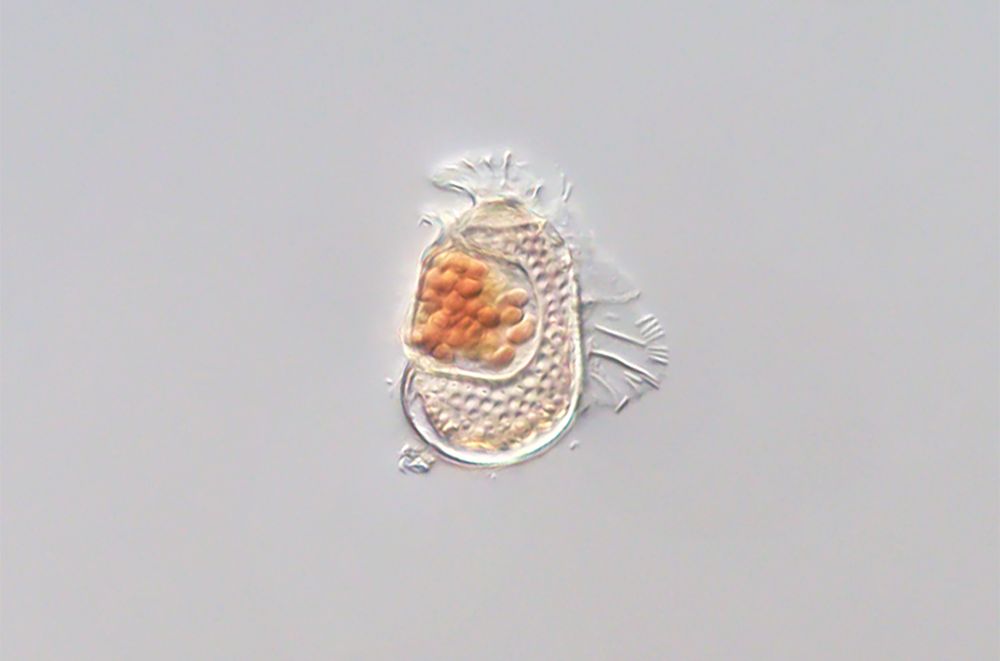


www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵

www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵

rdcu.be/eg4OA
1/
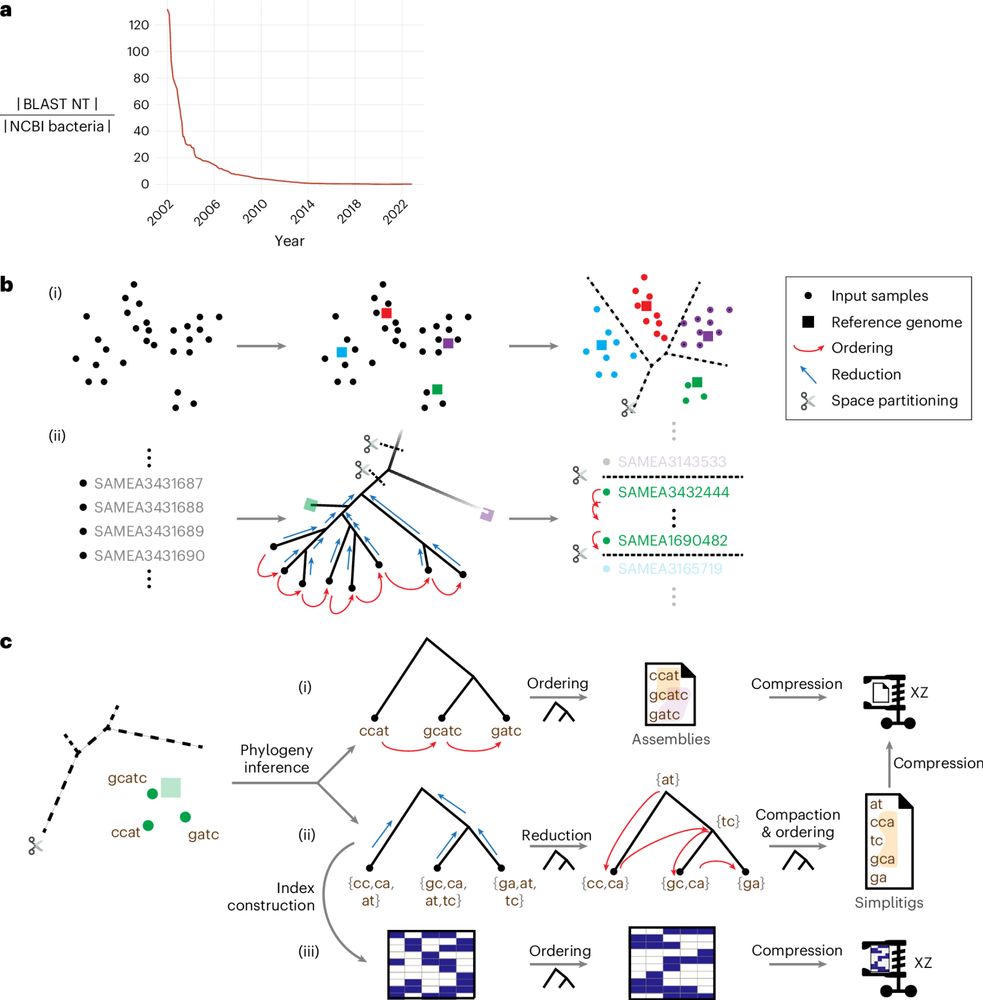
rdcu.be/eg4OA
1/
This was an early experiment to study how microbes spread through buildings and cause infections...

This was an early experiment to study how microbes spread through buildings and cause infections...
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)



#nanopore #basecaller #syntheticbiology

#nanopore #basecaller #syntheticbiology