
https://nmr.chemie.uni-koeln.de/
Explore the power of #NMR spectroscopy and its impactful applications in #structuralbiology, #drugdiscovery, and #metabolomics here at CERM/CIRMMP!
Register now! events.teams.microsoft.com/event/d9ac1b...

Explore the power of #NMR spectroscopy and its impactful applications in #structuralbiology, #drugdiscovery, and #metabolomics here at CERM/CIRMMP!
Register now! events.teams.microsoft.com/event/d9ac1b...
Our latest paper explores how small molecules can bind SARS-CoV-2 RNA, combining in silico screening with NMR experiments to uncover promising RNA-targeting candidates.
full text: chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/...
#RNA #NMR #screening #drugtargeting

Our latest paper explores how small molecules can bind SARS-CoV-2 RNA, combining in silico screening with NMR experiments to uncover promising RNA-targeting candidates.
full text: chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/...
#RNA #NMR #screening #drugtargeting




NMR spectroscopy studies of hydrogen bonding
(Dračínský) – Coord. Chem. Rev.: doi.org/10.1016/j.cc...
@iocbprague.bsky.social @czechacademy.bsky.social

NMR spectroscopy studies of hydrogen bonding
(Dračínský) – Coord. Chem. Rev.: doi.org/10.1016/j.cc...
@iocbprague.bsky.social @czechacademy.bsky.social
doi.org/10.1038/s414...

doi.org/10.1038/s414...

The Etzkorn Lab is looking for up to 7 highly motivated PostDocs and 1-2 PhD candidates to support our endeavour to translate the DNAzyme technology into therapeutic applications. For more information see: www.ipb.hhu.de/fileadmin/re...
Please repost. #Postdoc, @hhu.de

Full text: academic.oup.com/nar/article/...
#G4 #G-Quadruplex #NMR #smFRET #ChIP-qPCR

Full text: academic.oup.com/nar/article/...
#G4 #G-Quadruplex #NMR #smFRET #ChIP-qPCR


👉 Synthesis and proof of principle is here: chemistry-europe.onlinelibrary.wiley.com/doi/full/10....
2/n

👉 Synthesis and proof of principle is here: chemistry-europe.onlinelibrary.wiley.com/doi/full/10....
2/n
tldr: Apple used flow matching models to more efficiently predict protein folding structures.
More of this, please... from ALL big tech.

tldr: Apple used flow matching models to more efficiently predict protein folding structures.
More of this, please... from ALL big tech.
This #discovery was made by a research team @bccologne.bsky.social studying the molecular architecture of #synapses. The study shows that the protein #gephrin forms flexible filaments in the brain, serving as an important building block of inhibitory synapses.
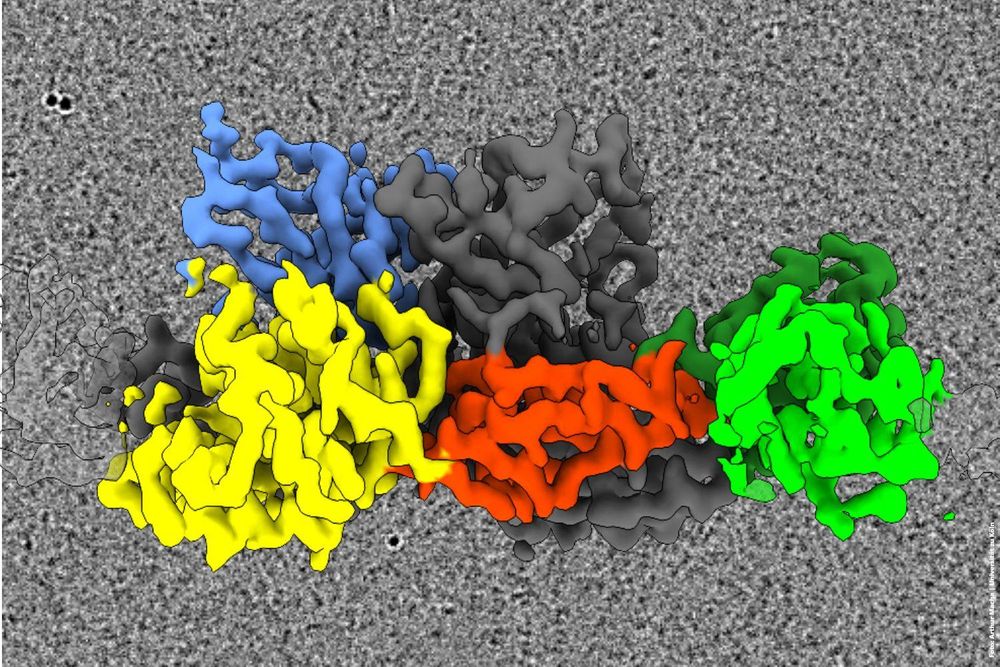
@instruct-eric.bsky.social @gdch.de
#NMR #EPR #FGMR #GDCh #InstructERIC


emploi.cnrs.fr/Offres/CDD/U... #NMRjobs #NMRchat 🧲
emploi.cnrs.fr/Offres/CDD/U... #NMRjobs #NMRchat 🧲
academic.oup.com/nar/article/...

academic.oup.com/nar/article/...

academic.oup.com/nar/article/...
Our selective arginine labeling yields well-resolved 1H-detected spectra in solids (and solution). We apply it to study dynamics in crystalline ubiquitin and a >130 kDa large enzyme
1/2
Our selective arginine labeling yields well-resolved 1H-detected spectra in solids (and solution). We apply it to study dynamics in crystalline ubiquitin and a >130 kDa large enzyme
1/2
www.nature.com/articles/s41...

We identified evolutionary origins of many fungal effectors!
We show that fungi secrete lots of antimicrobial proteins, and that some of them were repurposed by plant pathogens for host immune suppression.
www.biorxiv.org/content/10.1...
cc @teamthomma.bsky.social



pubs.acs.org/doi/10.1021/...

pubs.acs.org/doi/10.1021/...
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
go.nature.com/4lSCdzE
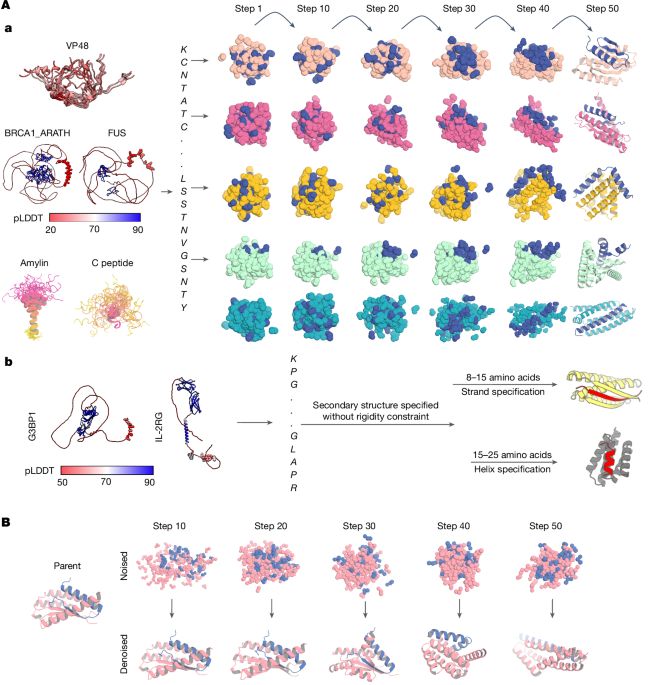
go.nature.com/4lSCdzE

