ML for Small-Molecule Binding Protein Design
Polizzi Lab at Dana Farber Cancer Institute
🏳️🌈


Can protein language models help us fight viral outbreaks? Not yet. Here’s why 🧵👇
1/12

Can protein language models help us fight viral outbreaks? Not yet. Here’s why 🧵👇
1/12




and the preprint www.biorxiv.org/content/10.1...
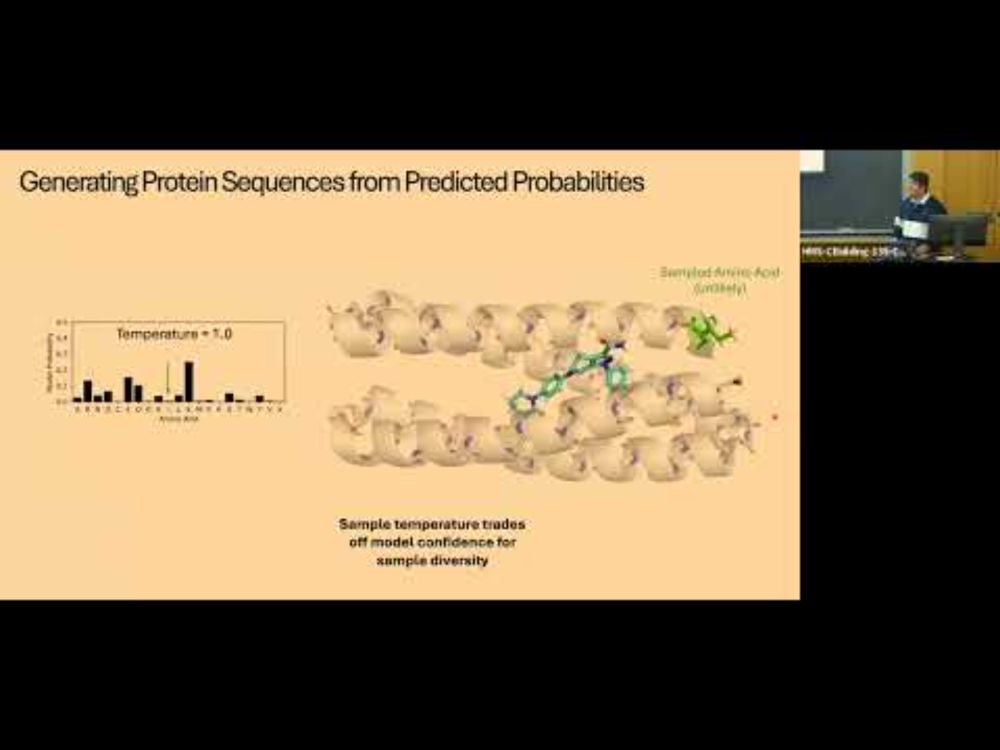
and the preprint www.biorxiv.org/content/10.1...




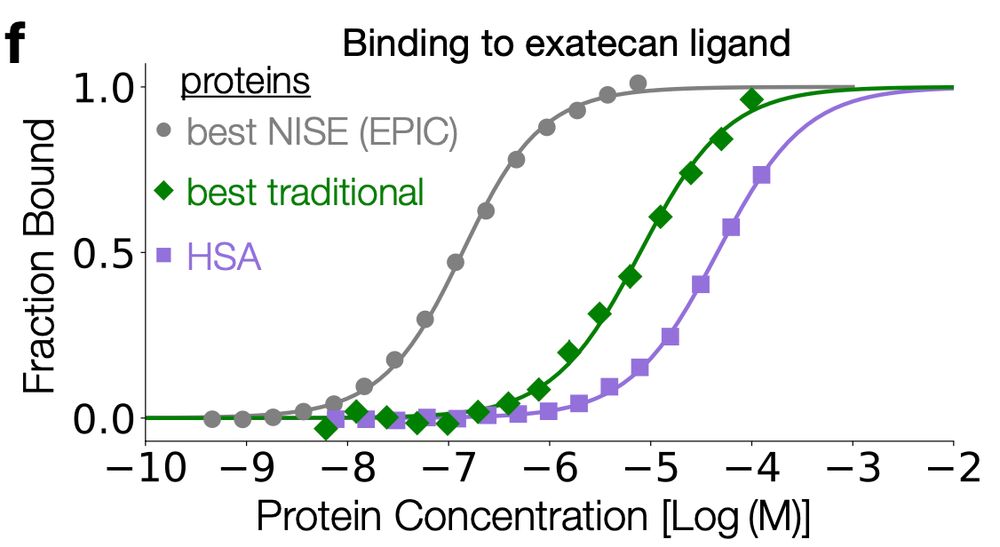
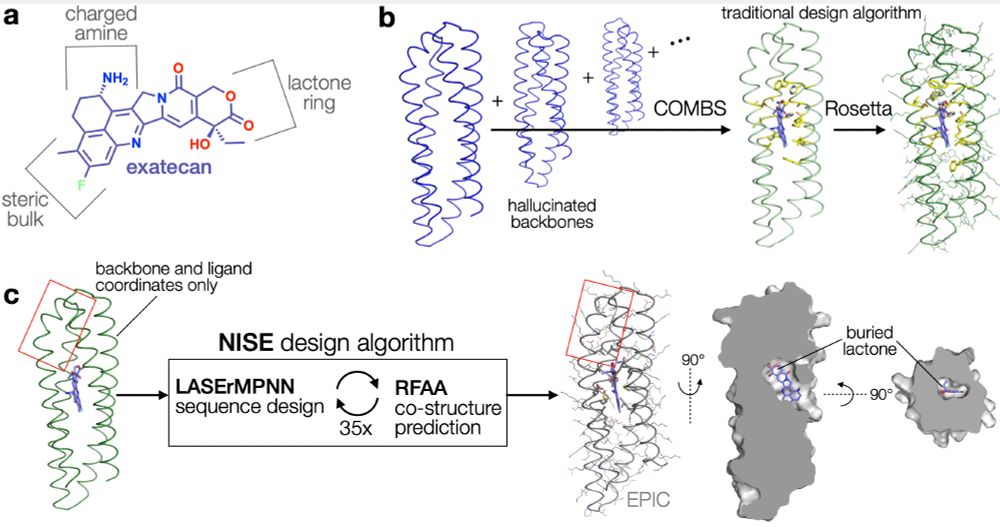
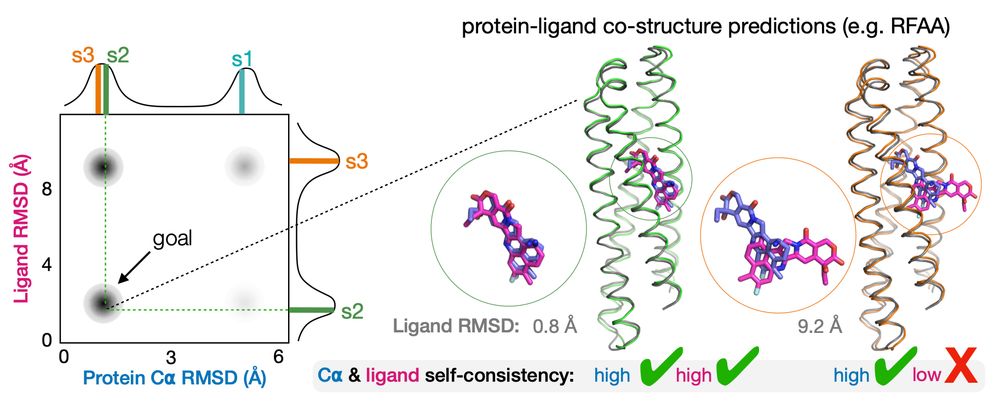
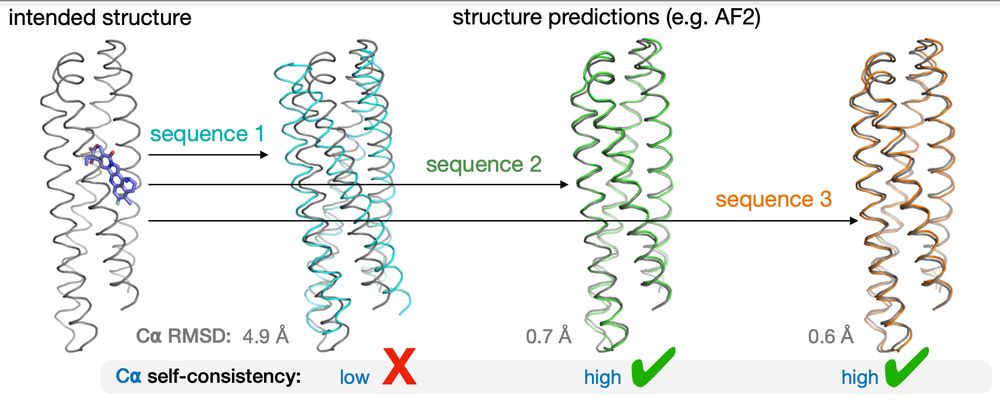
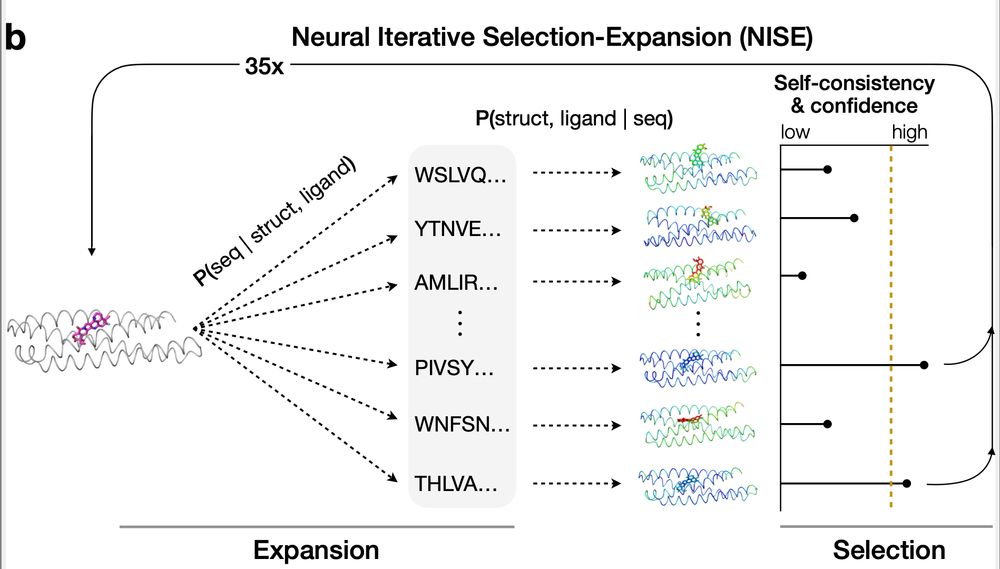
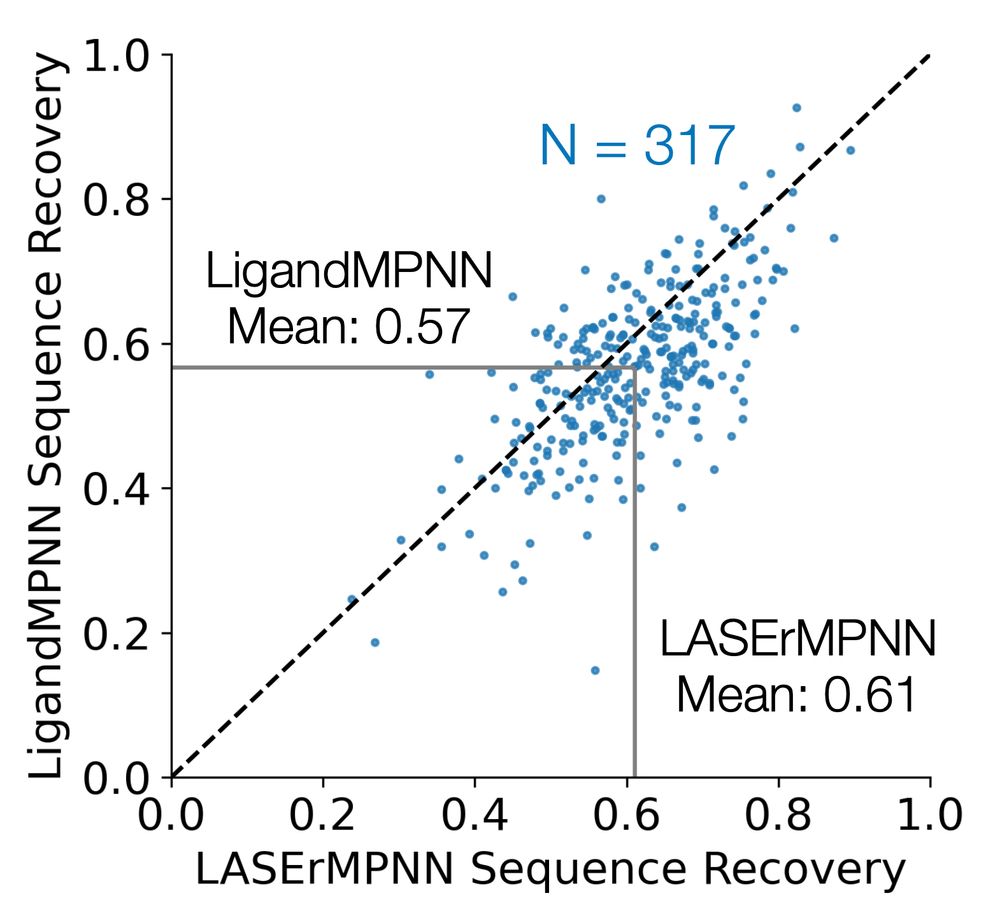
🧵🧪

🧵🧪

