Daniel Wendscheck
@dwendscheck.bsky.social
790 followers
1K following
71 posts
PhD student (close to finish!) at the University Würzburg. Likes to play with structure prediction tools, Crosslinking-MS, Native MS and bicycles. Proud of DOI:10.1073/pnas.2009502117 and DOI:10.1101/2024.04.30.591961 #TeamMassSpec #phdlife #peroxisomes
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Eddie Rashan
@edreesrashan.bsky.social
· Jun 22

ACAD10 and ACAD11 enable mammalian 4-hydroxy acid lipid catabolism - Nature Structural & Molecular Biology
Rashan, Bartlett and colleagues show that mammalian 4-hydroxy fatty acids are primarily catabolized by ACAD10 and ACAD11 (atypical mitochondrial and peroxisomal acyl-CoA dehydrogenases, respectively) ...
doi.org
Reposted by Daniel Wendscheck
Reposted by Daniel Wendscheck
Amrei Bahr
@amreibahr.bsky.social
· Jun 17
Keine Spitze ohne Berg: Zum Begriff des „Spitzenforschers“
Der Begriff des Spitzenforschers degradiert das Gros der Wissenschaftler_innen als zweitklassig und stützt einen Geniekult, der der Arbeitsrealität in der Wissenschaft weder angemessen noch zuträglich...
arbeitinderwissenschaft.substack.com
Reposted by Daniel Wendscheck
Amrei Bahr
@amreibahr.bsky.social
· Jun 16
Reposted by Daniel Wendscheck
Michael MacCoss
@maccoss.bsky.social
· Jun 16
Nature Methods
@natmethods.nature.com
· Jun 16
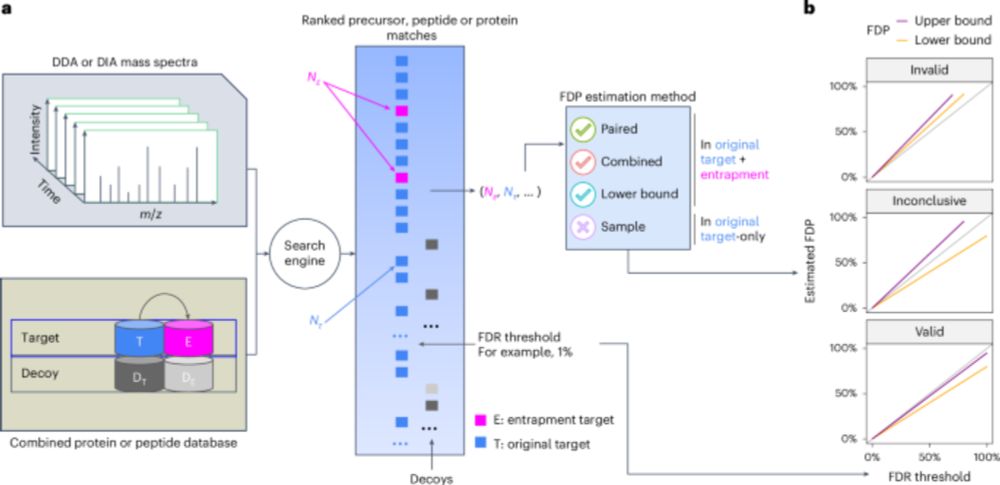
Assessment of false discovery rate control in tandem mass spectrometry analysis using entrapment - Nature Methods
A theoretical foundation for entrapment methods is presented, along with a method that enables more accurate evaluation of false discovery rate (FDR) control in proteomics mass spectrometry analysis p...
www.nature.com

















