BPOET: 1st high-throughput search (10,000 cmpds), resuscitates persisters (2020) with mechanism of activating hibernating ribosomes via 23S rRNA pseudouridine synthase RluD (E. coli, doi:10.1111/1462-2920.14828), cited, but not in context vs. new search:


by Thomas K. Wood — Reposted by Thomas K. Wood
Reposted by Thomas K. Wood
Nasal colonisation by S. aureus is linked with depression in a human cohort and shown in a mouse model to cause decreased serotonin and dopamine in the brain
#MicroSky #Depression
www.nature.com/articles/s41...

Reposted by Thomas K. Wood

Balanced review from Ed Dudley considering whether CRISPR-Cas is active in E. coli K-12. We discovered it silences lysis transcripts from cryptic prophages in 2022 (doi 10.3390/ijms232416195).

On the basis of sex: another important link of toxin/antitoxin systems to the stress response. Here, P. aeruginosa-derived NQNO inhibits N. gonorrhoeae by increasing oxidative stress, activating Zeta1 toxin, without affecting vaginal lactobacilli.

We found ubiquitous recombinases from cryptic prophages may invert DNA to form new, chimeric proteins that inhibit phages by blocking adsorption. Congratulations to Joy, Daniel, Rodolfo and Michael for their strong efforts.

(2/2) Not cited: doi:10.1111/1462-2920.14075 where it was shown the few live VBNCs = persisters. Also, there is no proof of differences in dormancy, i.e., no differences in persister cells (enviromicro-journals.onlinelibrary.wiley.com/doi/10.1111/...).

(1/2) Fantastic summary of flawed research. As published, there are no differences between persisters & the live fraction of VBNCs & one of the clear indications of death is protein aggregation, so increased aggregation simply means the cells are dead.
Reposted by Thomas K. Wood
in @narjournal.bsky.social by Lin et al.
academic.oup.com/nar/article/...
🧐106 claims of cell death ('Abi', 'PCD', 'abortosome', 'death') upon phage induction of Hailibu system (DUR4297 DNase + HerA ATPase), but extraordinarily weak data to support claims: just system fails 3 MOI but works at 100X less phage.

Chicken or egg? Looks like toxin/antitoxins were first, and a type III TA system (HEPN RNase toxin inhibited by ncRNA) evolved into CRISPR-Cas13 (RNA-degrading).

Excellent additional proof of the regulatory nature of toxin/antitoxin modules by binding promoters other than the ones that encode the TA locus. Here, MtvT/MvtA decrease conjugation and increase virulence of P. aeruginosa.

Results are in-line with the first single-cell persister resuscitation work of 2018 (not cited) which showed many persister cells lyse upon waking due to internalized ampicillin (doi 10.1111/1462-2920.14093).

🧐Yes, indeed, there is a continuum of dormancy: Persisters revive based on ribosomes almost instantaneously with food and VBNCs never revive since they are dead, which is the ultimate dormant state...i.e., protein aggregation = death.

Most 'novel' systems described here after prediction from machine learning, are related to toxin/antitoxin systems (e.g., DS-11, DS6AB, DS-3, DS-32, HEPN motifs, etc.) cementing the idea that TAs are the most prevalent phage defense system.

Yet another phage defense system that detects free DNA ends and provides immunity without causing 'programmed cell death'.

Reposted by Thomas K. Wood
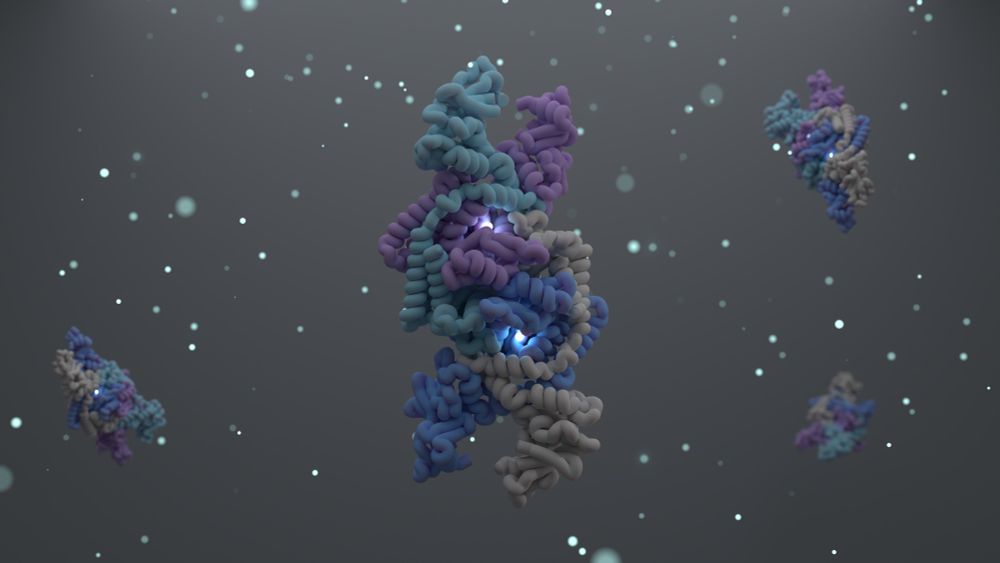
Today in @cellpress.bsky.social we report the structure and function of the Shedu anti-phage defense system.
tinyurl.com/4crj6dnx
A long 🧵...
Another phage inhibition system (Zorya) that does not cause cell death or what has been termed 'abortive infection'. Congratulations to the authors for discerning the lovely mechanism of detecting phage disruption of the cell envelope then DNase activity.
Just my opinion, based on what I consider sound research in a field with much noise due to studying dying cells, but please see the attached list of the usual errors from this lab in their Nature Rev Micro persister review.

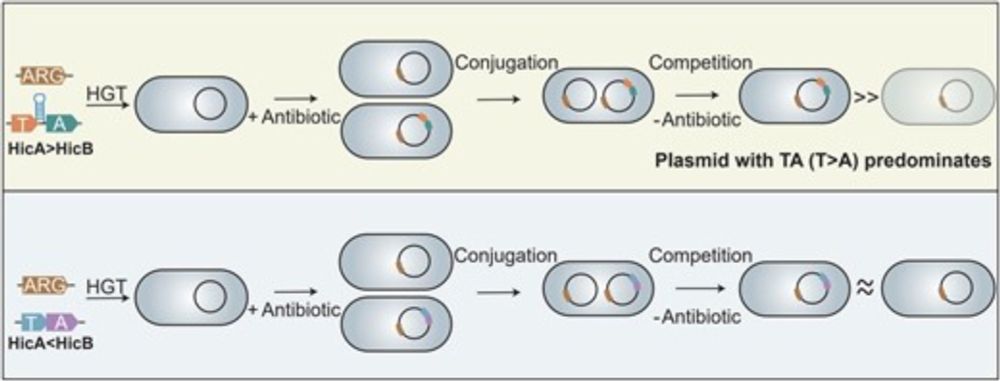
I'ing tripartite TA system identified in Salmonella Gifsy-1 prophage (not called this but toxin is common HEPN protein & TAs are prevalent in prophages). No survival phenotype upon TA deletion (Fig. 1M), but less lysis from other prophages (due to slower growth?).


