
Evolution | Population genetics | Plant-microbe interactions
https://yusugihara.github.io/
The helper NLR NRC3 evolved to evade inhibition by the cyst nematode effector SS15 over 19 million years ago.
Check out the peer-reviewed version of our preprint:
🔗 doi.org/10.1371/jour...

The helper NLR NRC3 evolved to evade inhibition by the cyst nematode effector SS15 over 19 million years ago.
Check out the peer-reviewed version of our preprint:
🔗 doi.org/10.1371/jour...


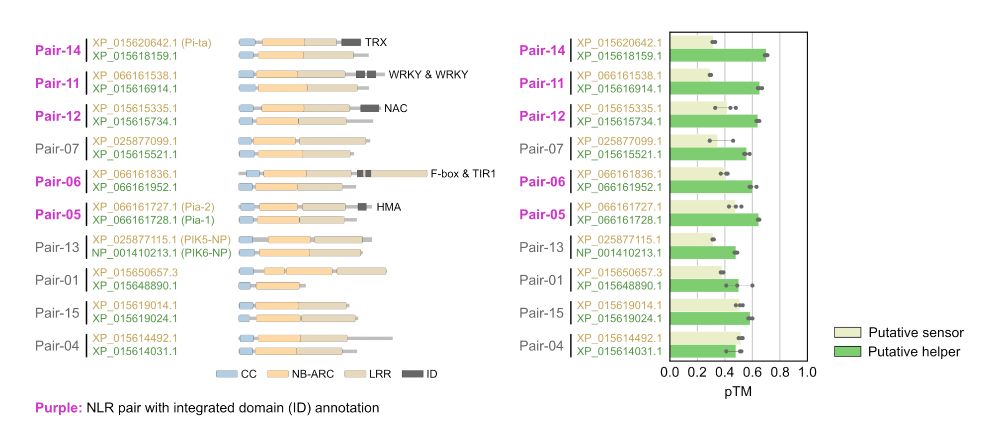


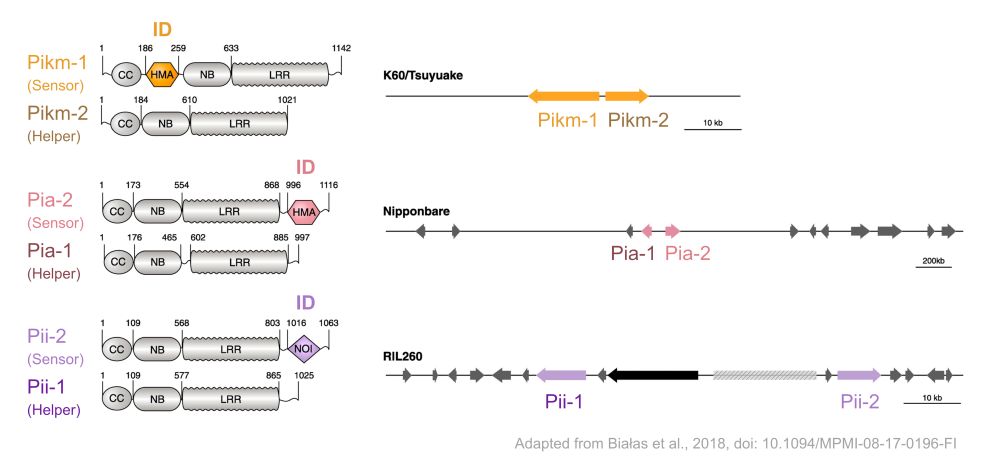
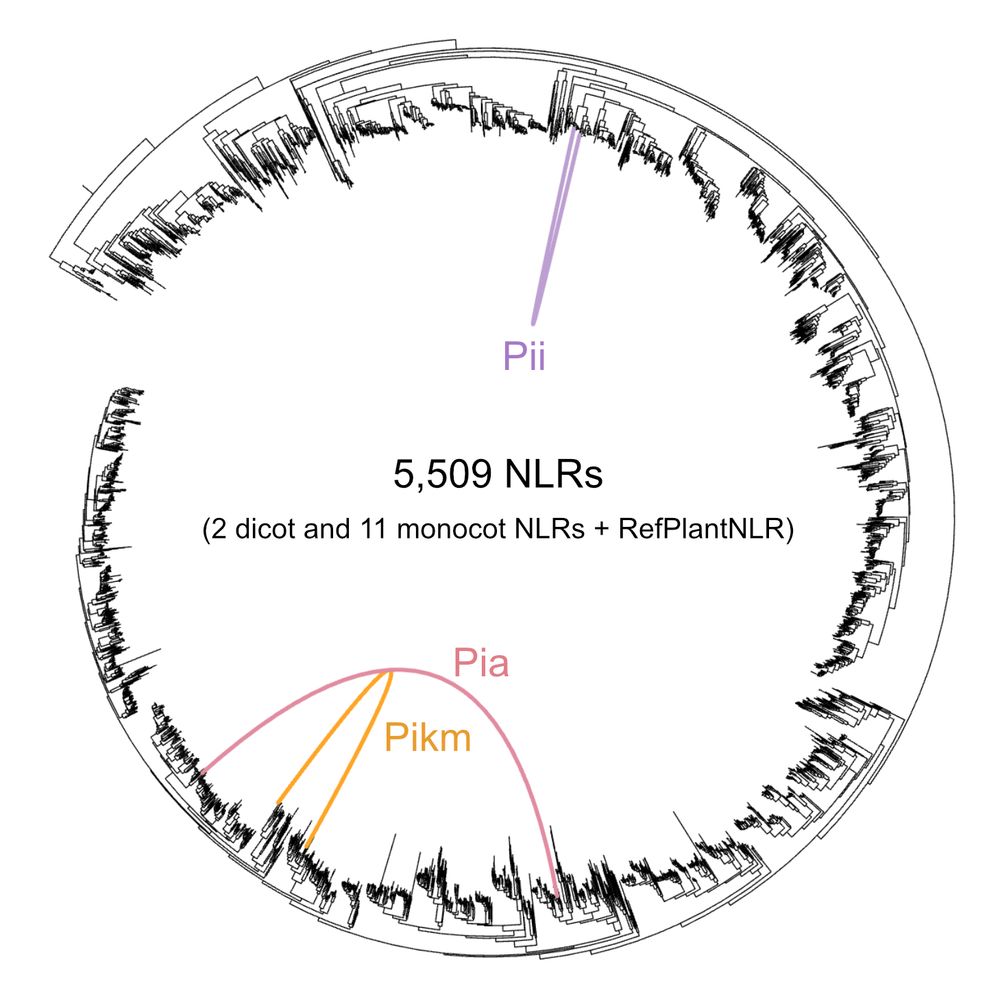
doi.org/10.1101/2024...
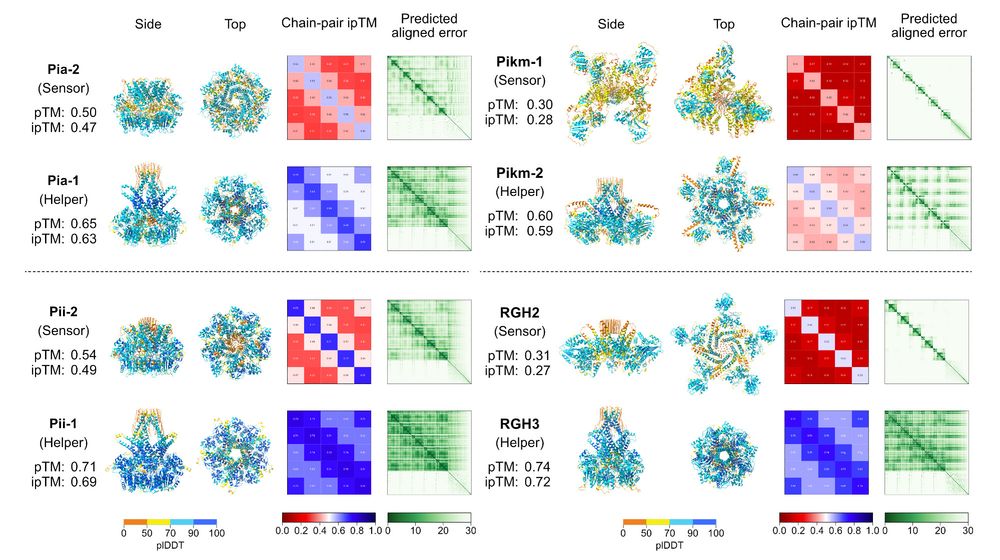
doi.org/10.1101/2024...

