
«Ara mateix / enfilo aquesta agulla amb el fil d'un propòsit que no dic / i em poso a apedaçar.»
xgrau.github.io & ecoevo.social/@xgrau
This looks great. Figure 2 is textbook material!
www.nature.com/articles/s41...

This looks great. Figure 2 is textbook material!
www.nature.com/articles/s41...
sebelab.crg.eu/multicoral-s...

sebelab.crg.eu/multicoral-s...

🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...


🪸 🌊
#evobio #corals #coralbiology
www.nature.com/articles/s41...
(poster P01.286 in the symposium on «Evolutionary Genomics: Understanding & Adapting to #ClimateChange», which is looking great! 🙂)

(poster P01.286 in the symposium on «Evolutionary Genomics: Understanding & Adapting to #ClimateChange», which is looking great! 🙂)


«Chromatin loops are an ancestral hallmark of the animal regulatory genome»
It’s been a pleasure and a privilege to have been able to contribute to @ianakim.bsky.social’s project 🙂 — check out her thread below!


«Chromatin loops are an ancestral hallmark of the animal regulatory genome»
It’s been a pleasure and a privilege to have been able to contribute to @ianakim.bsky.social’s project 🙂 — check out her thread below!
Here @crisnava.bsky.social, @seanamontgomery.bsky.social & collaborators develop a novel ChIPseq protocol, and demonstrate its huge potential to study the evolution of chromatin function and regulation across the eukaryotic tree of life.

Here @crisnava.bsky.social, @seanamontgomery.bsky.social & collaborators develop a novel ChIPseq protocol, and demonstrate its huge potential to study the evolution of chromatin function and regulation across the eukaryotic tree of life.
Open positions:
1. Looking for a developer for the BCA database, with @ebi.embl.org
2. A senior research technician to develop single-cell methods
3. A bioinformatician to analyse new atlases, with @sangerinstitute.bsky.social
Info in next post.

Open positions:
1. Looking for a developer for the BCA database, with @ebi.embl.org
2. A senior research technician to develop single-cell methods
3. A bioinformatician to analyse new atlases, with @sangerinstitute.bsky.social
Info in next post.
ouyanglab.com/singlecell/b...). My only (very minor) objection would be about heatmaps in general (we put scales in them but often read them in a qualitative way), but the layout of the clusters does a good job at enabling comparison of differential marker expression.

ouyanglab.com/singlecell/b...). My only (very minor) objection would be about heatmaps in general (we put scales in them but often read them in a qualitative way), but the layout of the clusters does a good job at enabling comparison of differential marker expression.
For this project we’re going go work with the lovely sea anemome, Nematostella ❤️
We’ll combine population genomics with single-cell omics to build whole-organism genotype-phenotype maps & probe the genetic determinants of cell type evolution.

For this project we’re going go work with the lovely sea anemome, Nematostella ❤️
We’ll combine population genomics with single-cell omics to build whole-organism genotype-phenotype maps & probe the genetic determinants of cell type evolution.
I haven’t reread it since, weary of finding too many aspects in which I might have been too naïve… maybe it’s time to embrace this change though!

I haven’t reread it since, weary of finding too many aspects in which I might have been too naïve… maybe it’s time to embrace this change though!
«GeneExt: a gene model extension tool for enhanced single-cell RNA-seq analysis»
www.biorxiv.org/content/10.1...
Presented in this thread by Grisha Zolotarov:
twitter.com/zolotarg/sta...
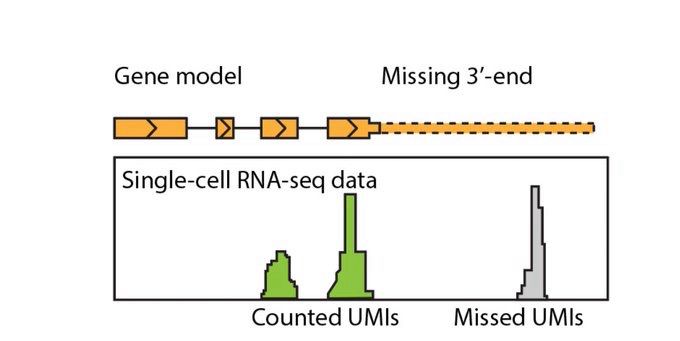
«GeneExt: a gene model extension tool for enhanced single-cell RNA-seq analysis»
www.biorxiv.org/content/10.1...
Presented in this thread by Grisha Zolotarov:
twitter.com/zolotarg/sta...


