
www.nature.com/articles/s41...
With Gillian Rodger, @nstoesser.bsky.social, @samlipworth.bsky.social, @stat-sarah.bsky.social, and many others!
www.nature.com/articles/s41...
With Gillian Rodger, @nstoesser.bsky.social, @samlipworth.bsky.social, @stat-sarah.bsky.social, and many others!
Thread 1/n

Thread 1/n
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
Nanopore's getting accurate, but
1. Can this lead to better metagenome assemblies?
2. How, algorithmically, to leverage them?
with co-author Max Marin @mgmarin.bsky.social, supervised by Heng Li @lh3lh3.bsky.social
1 / N
📌Community outbreak of OXA-48–producing Escherichia coli linked to food premises, New Zealand, 2018-2022
doi.org/10.3201/eid3...
Thanks to folks across NZ who helped investigate & respond
#AMR #PublicHealth #Genomics #Epidemiology #Scicomm
🧵
🖥️🧬💻
#AcademicSky
#MicroSky
🧪🧫🦠

📌Community outbreak of OXA-48–producing Escherichia coli linked to food premises, New Zealand, 2018-2022
doi.org/10.3201/eid3...
Thanks to folks across NZ who helped investigate & respond
#AMR #PublicHealth #Genomics #Epidemiology #Scicomm
🧵
🖥️🧬💻
#AcademicSky
#MicroSky
🧪🧫🦠
Don't miss the opportunity to be a part of this exciting conference on microbial typing & #genomics. The theme this year, Microbial typing: from fundamental to daily microbiology
Register today and see you in Porto!
www.escmid.org/congress-eve...

Don't miss the opportunity to be a part of this exciting conference on microbial typing & #genomics. The theme this year, Microbial typing: from fundamental to daily microbiology
Register today and see you in Porto!
www.escmid.org/congress-eve...
🧫 Automated antibiograms
💻 Integration to WHONET
🧪 Updated CLSI and EUCAST breakpoints
💡 Free
amr-for-r.org
#biostatistics #episky #Statistics #AMR
🧫 Automated antibiograms
💻 Integration to WHONET
🧪 Updated CLSI and EUCAST breakpoints
💡 Free
amr-for-r.org
#biostatistics #episky #Statistics #AMR
In it, I benchmark the new version of Dorado from @nanoporetech.com, which comes with new DNA basecalling models. Short version: big accuracy gains for hac, small improvements for sup.
Check it out for the full results:
rrwick.github.io/2025/05/27/d...
In it, I benchmark the new version of Dorado from @nanoporetech.com, which comes with new DNA basecalling models. Short version: big accuracy gains for hac, small improvements for sup.
Check it out for the full results:
rrwick.github.io/2025/05/27/d...
www.biorxiv.org/content/10.1...
(1/6)

www.biorxiv.org/content/10.1...
(1/6)
Autocycler, the automated successor to Trycycler from @rrwick.bsky.social has a pre-print out - overall, pretty awesome performance (and is very easy to use)
github.com/rrwick/Autoc...

Autocycler, the automated successor to Trycycler from @rrwick.bsky.social has a pre-print out - overall, pretty awesome performance (and is very easy to use)
github.com/rrwick/Autoc...
www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵

www.biorxiv.org/content/10.1...
and tell you just a little bit about it in the following 🧵
rdcu.be/eg4OA
1/
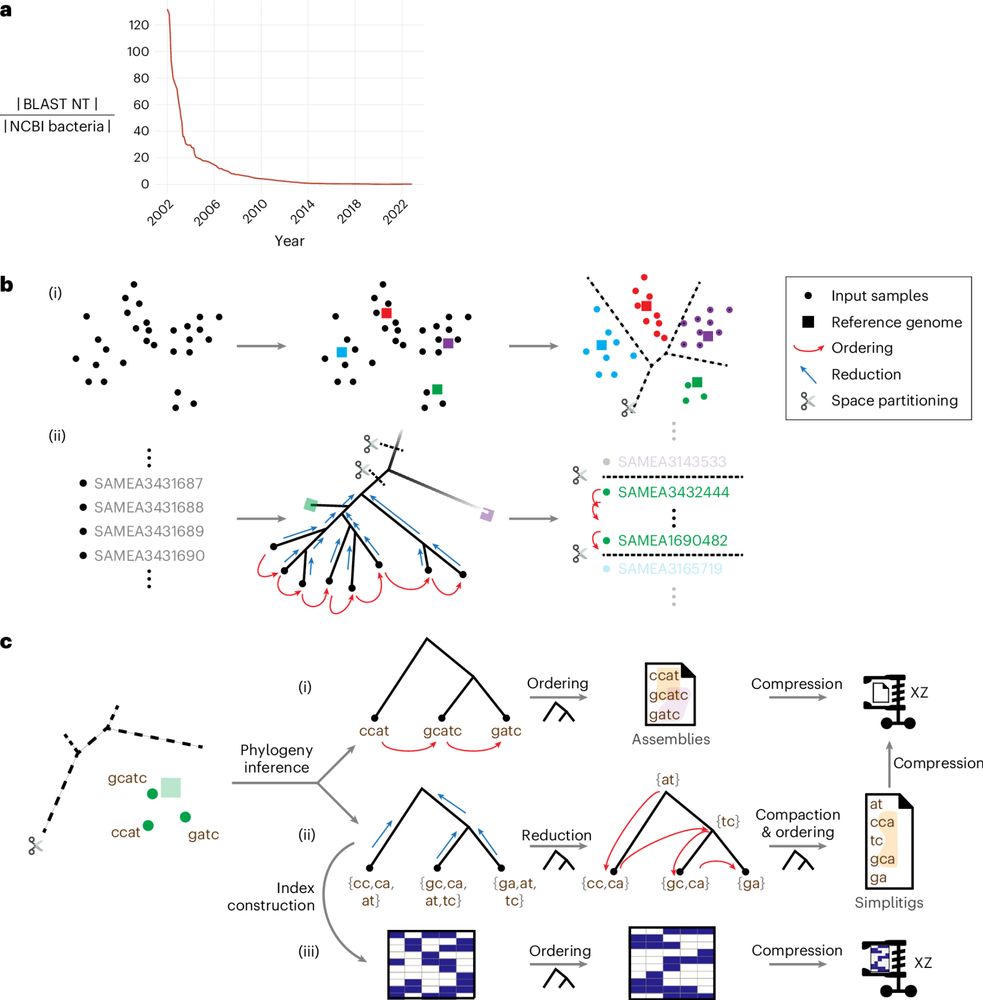
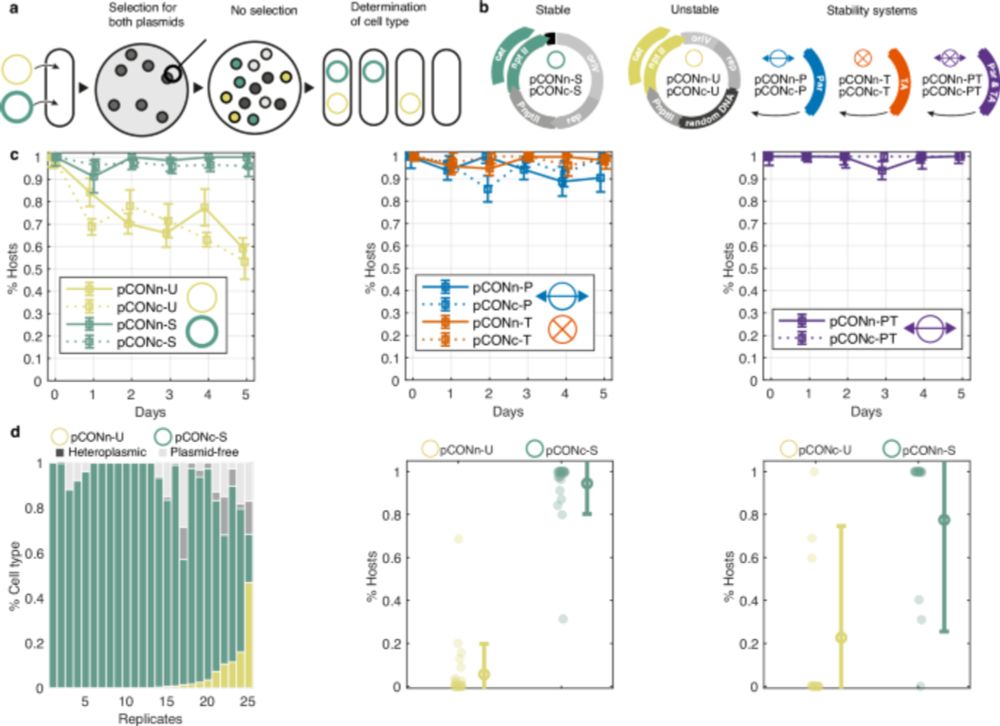
Our paper in @naturemethods.bsky.social answers this: use evolutionary history to guide compression and search.
rdcu.be/eg4OA
w/ @baym.lol, @zaminiqbal.bsky.social et al. 🧵1/

Our paper in @naturemethods.bsky.social answers this: use evolutionary history to guide compression and search.
rdcu.be/eg4OA
w/ @baym.lol, @zaminiqbal.bsky.social et al. 🧵1/

Under the hood it's the same BLAST executable and databases provided by NCBI, with a replica of the NIH's interface - providing an alternative to the US gov service that's less congested, faster & more reliable
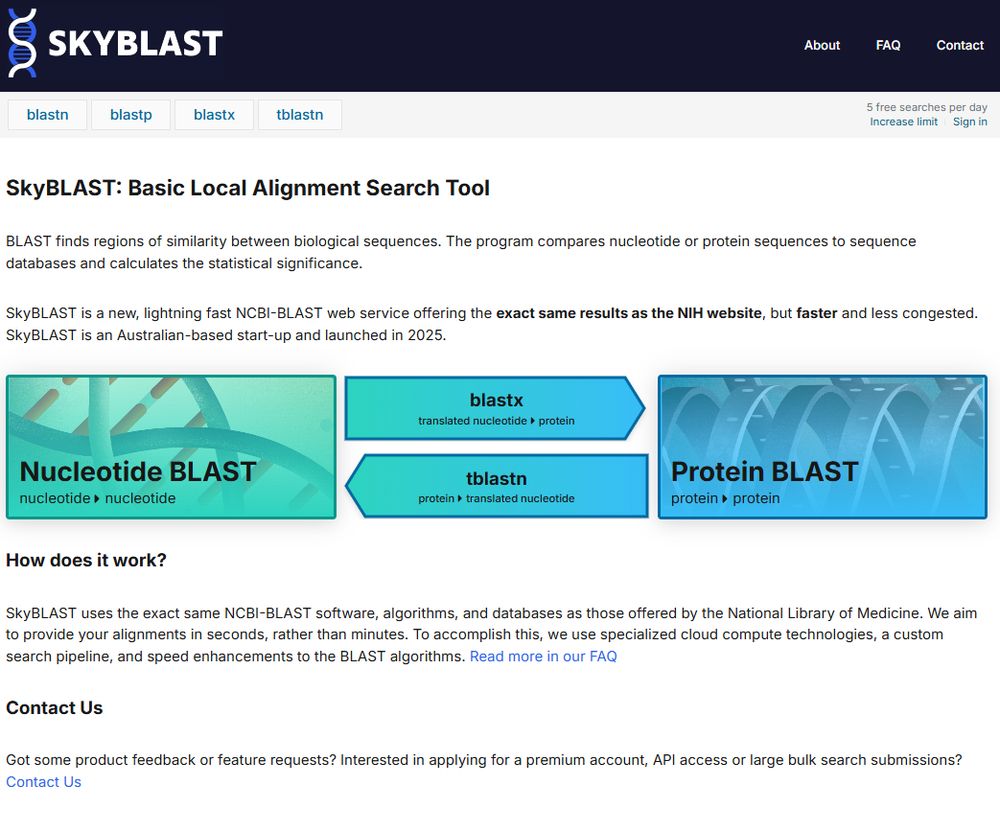
forms.gle/YJP6WQjsk8KK...

forms.gle/YJP6WQjsk8KK...
We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)

We investigate how well you can call variants directly from genome assemblies compared to traditional read-based variant calling.
Read it here: www.biorxiv.org/content/10.1...
Data & code: github.com/rrwick/Are-r...
(1/8)
Truly brilliant paper with great figures! @lancetmicrobe.bsky.social
www.thelancet.com/journals/lan...
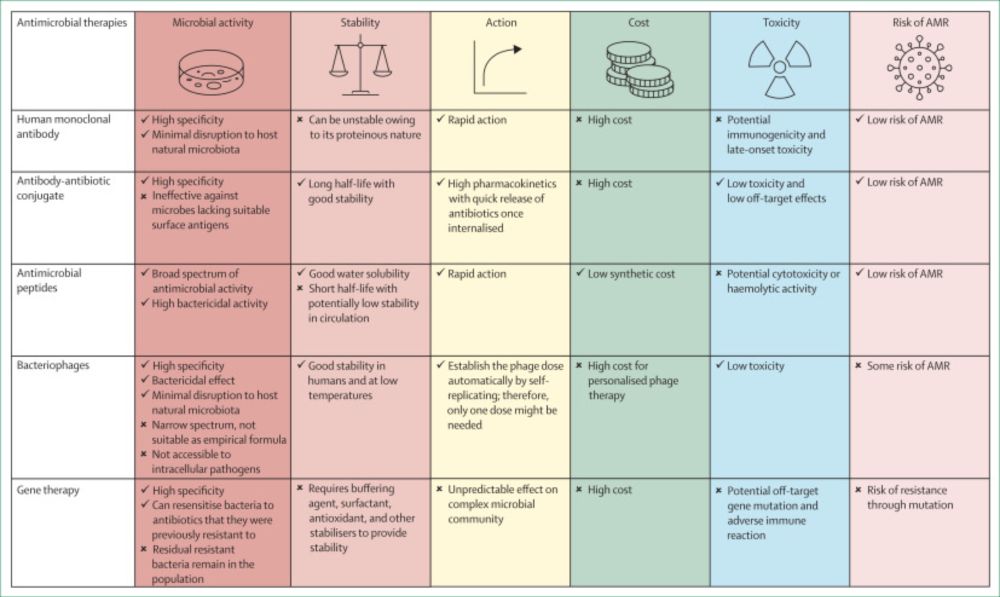
Truly brilliant paper with great figures! @lancetmicrobe.bsky.social
www.thelancet.com/journals/lan...

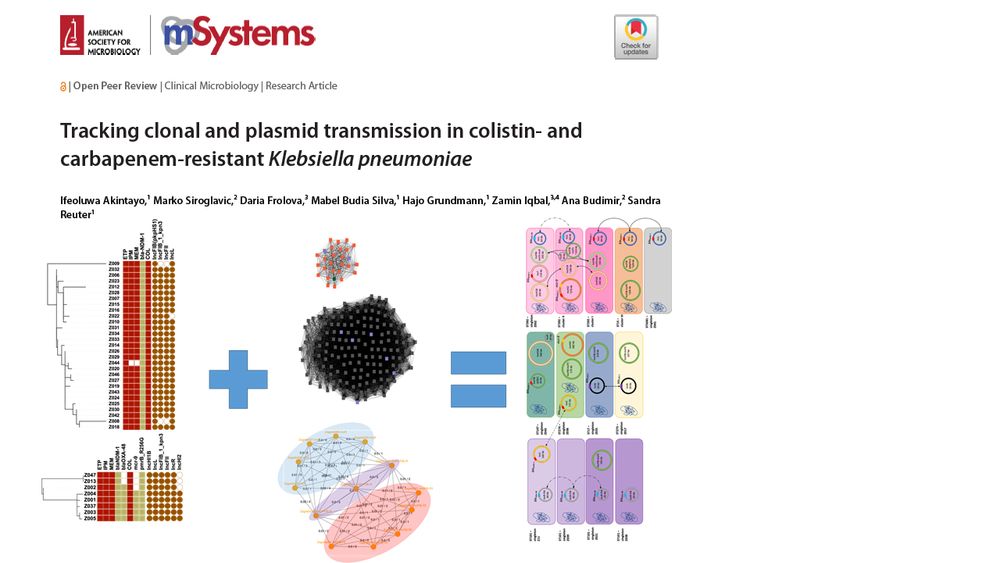
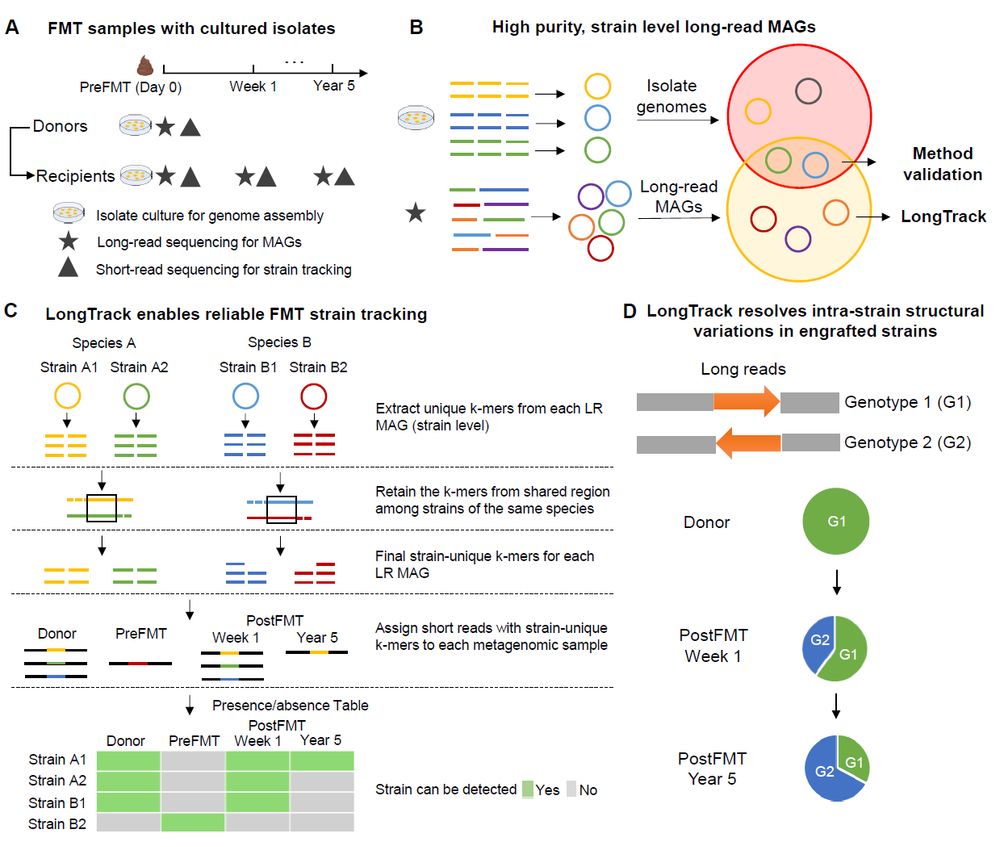
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)
I'm excited to announce Autocycler, my new tool for consensus assembly of long-read bacterial genomes!
It's the successor to Trycycler, designed to be faster and less reliant on user intervention.
Check it out: github.com/rrwick/Autoc...
(1/5)

