Algorithms, Machine Learning, Data Resources
Computational Genomics, Drug Discovery, Animal Tracking, etc.
wheelerlab.org
It just occurred to me: it's got to be cheaper to acquire a finished copy of your genome (high accuracy, phased, SV-resolved) than to print it as a collection of bound books.
(back-of-envelope checks out - anyone done the math?)

It just occurred to me: it's got to be cheaper to acquire a finished copy of your genome (high accuracy, phased, SV-resolved) than to print it as a collection of bound books.
(back-of-envelope checks out - anyone done the math?)


Simpatico: www.biorxiv.org/content/10.1...
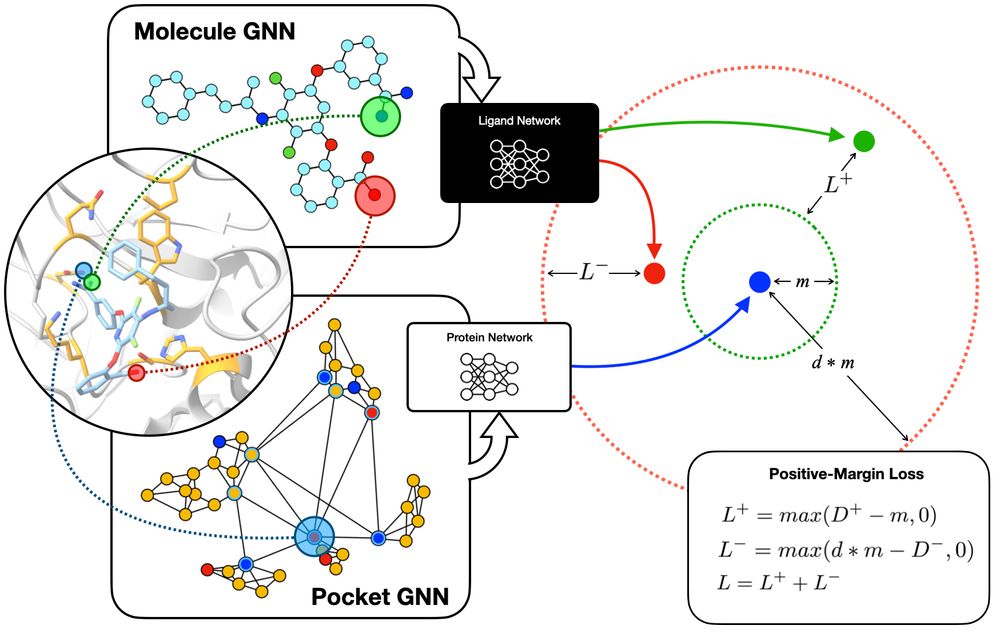
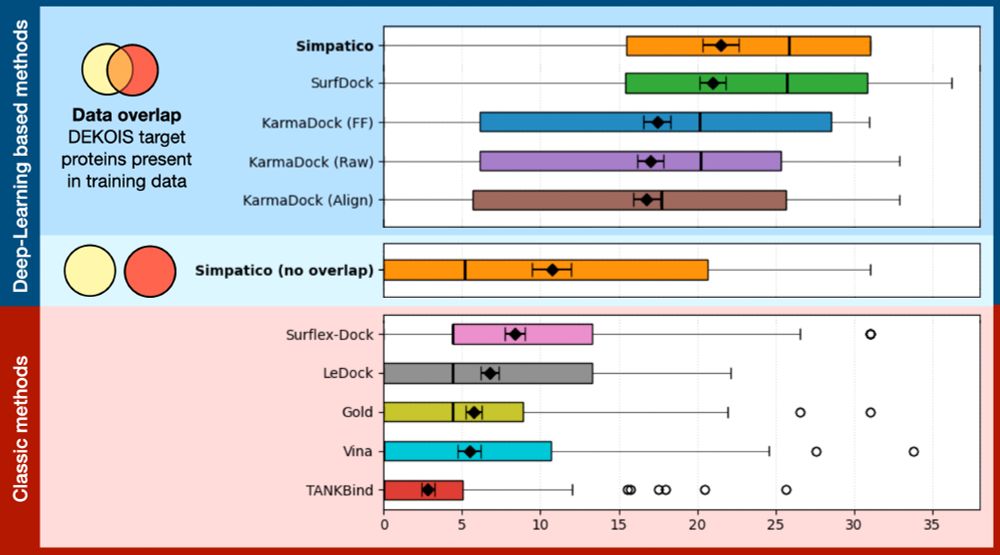
Simpatico: www.biorxiv.org/content/10.1...



www.reaganlibrary.gov/archives/spe...

www.reaganlibrary.gov/archives/spe...




So now we know how we should feel ...

So now we know how we should feel ...





“AI is dull as cold bread.”
But the bots, full of sass,
Said, “You’d better not pass,
Till you’ve laughed at the rhymes in your head!”
(This make me nervous)

“AI is dull as cold bread.”
But the bots, full of sass,
Said, “You’d better not pass,
Till you’ve laughed at the rhymes in your head!”
(This make me nervous)
github.com/TravisWheele...

github.com/TravisWheele...
This shows runtime vs recall on a test with ~1400 query MSAs, with ~20 true matches per query planted among 2M decoy sequences. Speed gains over H3 grow for bigger datasets, and scale linearly with MMSeqs2 runtime.

This shows runtime vs recall on a test with ~1400 query MSAs, with ~20 true matches per query planted among 2M decoy sequences. Speed gains over H3 grow for bigger datasets, and scale linearly with MMSeqs2 runtime.