We’ve added a few new analyses. First off, we show that, while gene presence absence variation (PAV) scales with evolutionary distance in both plants and animals, the base level and rate of accrual are both twice as high in plants.
We’ve added a few new analyses. First off, we show that, while gene presence absence variation (PAV) scales with evolutionary distance in both plants and animals, the base level and rate of accrual are both twice as high in plants.

Develop, benchmark, and apply Plant Genomic Language Models (gLMs) 🌿🧬💻
jobs.lbl.gov/jobs/computa...
#AI #PlantGenomics #gLMs #Postdoc

We explore the causes of this in soybean and cotton datasets in our recent preprint: www.biorxiv.org/content/10.1...
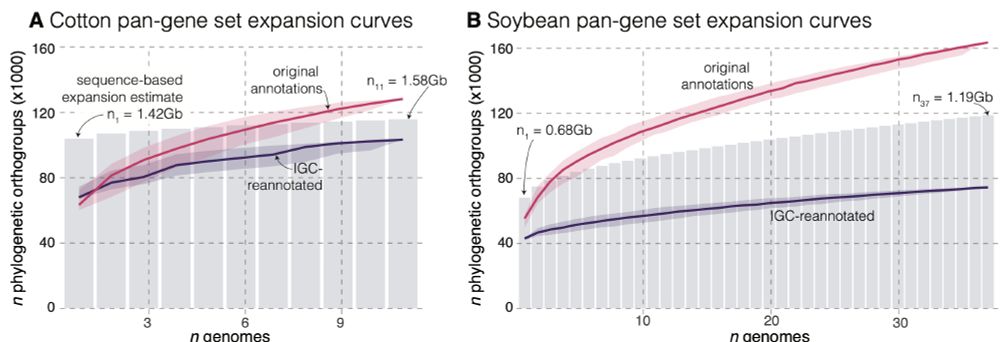
We explore the causes of this in soybean and cotton datasets in our recent preprint: www.biorxiv.org/content/10.1...

