
PhD, @bussilab.bsky.social SISSA |
#machinelearning, #RNA, #Proteins, #moleculardynamics matching experiments

Read our piece here: www.nature.com/articles/s43...
And the research article here: www.nature.com/articles/s43...
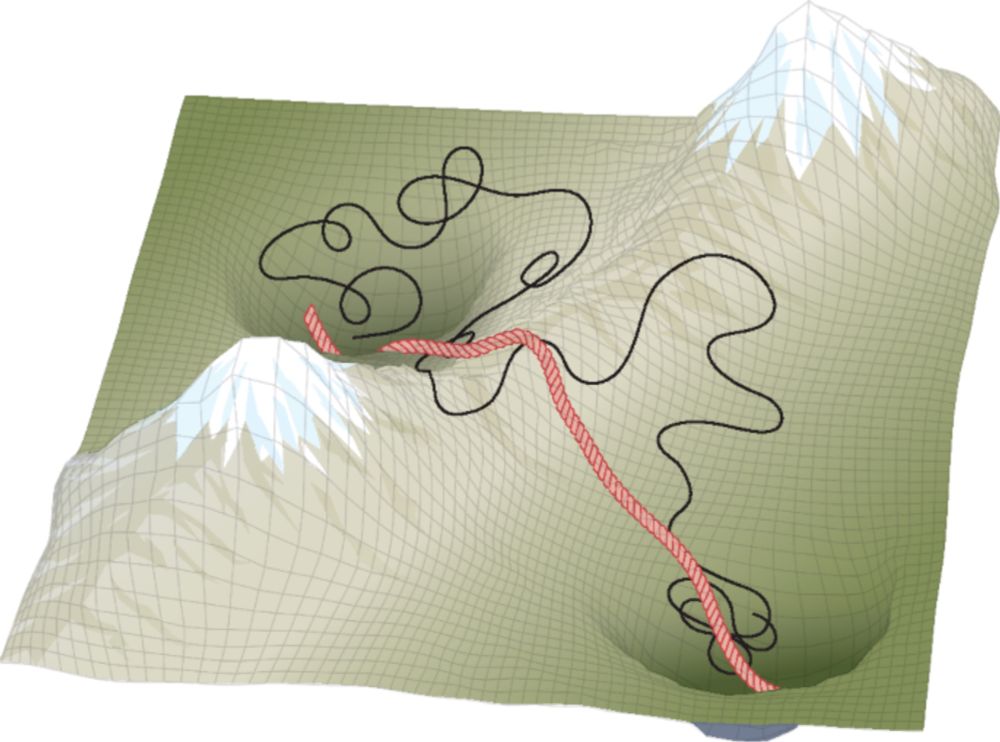
Read our piece here: www.nature.com/articles/s43...
And the research article here: www.nature.com/articles/s43...


