




▶️Concept suggested by Polach & Widom, 1996: www.sciencedirect.com/science/arti...
▶️Our first single-bp resolution model, 2010: www.cell.com/biophysj/ful...
▶️Its application to TFs at enhancers, 2011: iopscience.iop.org/article/10.1...

▶️Concept suggested by Polach & Widom, 1996: www.sciencedirect.com/science/arti...
▶️Our first single-bp resolution model, 2010: www.cell.com/biophysj/ful...
▶️Its application to TFs at enhancers, 2011: iopscience.iop.org/article/10.1...

▶️spatially integrates sequences and localises them relative to the target locus
▶️infers effective TF concentrations
▶️supplements with pairwise TF interactions

▶️spatially integrates sequences and localises them relative to the target locus
▶️infers effective TF concentrations
▶️supplements with pairwise TF interactions
project.iith.ac.in/sharmaglab/g...
addons.mozilla.org/en-US/firefo...

project.iith.ac.in/sharmaglab/g...
addons.mozilla.org/en-US/firefo...




▶️PU.1 can form strongly phased nucleosome arrays similar to CTCF

▶️PU.1 can form strongly phased nucleosome arrays similar to CTCF









▶️HMGB1 sites are away from where H1 binds
▶️HMGB1 increases nucleosomal DNA accessibility without displacing H1

▶️HMGB1 sites are away from where H1 binds
▶️HMGB1 increases nucleosomal DNA accessibility without displacing H1
▶️RoboATAC - automated ATAC-seq
▶️Deep learning models: motif orientation, spacing, flanking bases
▶️Nucleosome positioning analysis
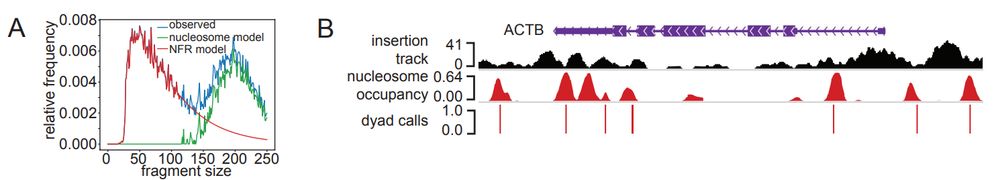
▶️RoboATAC - automated ATAC-seq
▶️Deep learning models: motif orientation, spacing, flanking bases
▶️Nucleosome positioning analysis


▶️Condensed purified native mononucleosomes with polyamines
▶️Nucleosomes from A compartments have low condensability and those from B compartments have high condensability
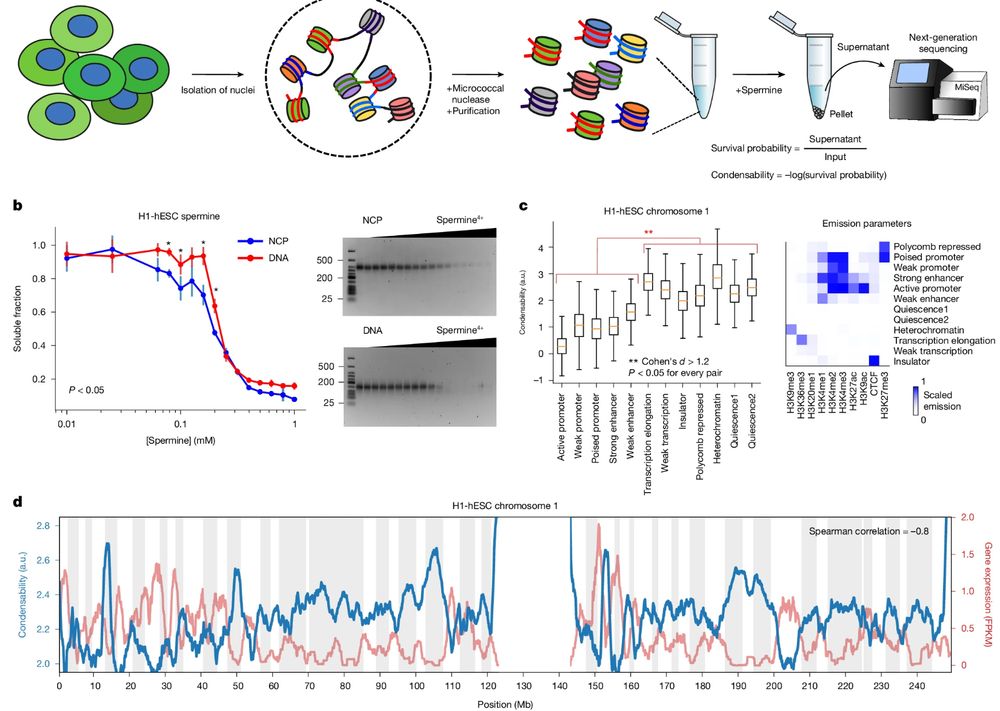
▶️Condensed purified native mononucleosomes with polyamines
▶️Nucleosomes from A compartments have low condensability and those from B compartments have high condensability
▶️In mature retina cells, the fraction of short-linker nucleosomes is lower -> stronger chromatin compaction.

▶️In mature retina cells, the fraction of short-linker nucleosomes is lower -> stronger chromatin compaction.
▶️FoxP3 recognizes a variety of TnG repeat microsatellites
▶️FoxP3 multimers are ultrastable, can bridge 2–4 DNA duplexes
▶️Nucleosomes can facilitate FoxP3 assembly by inducing local DNA bending
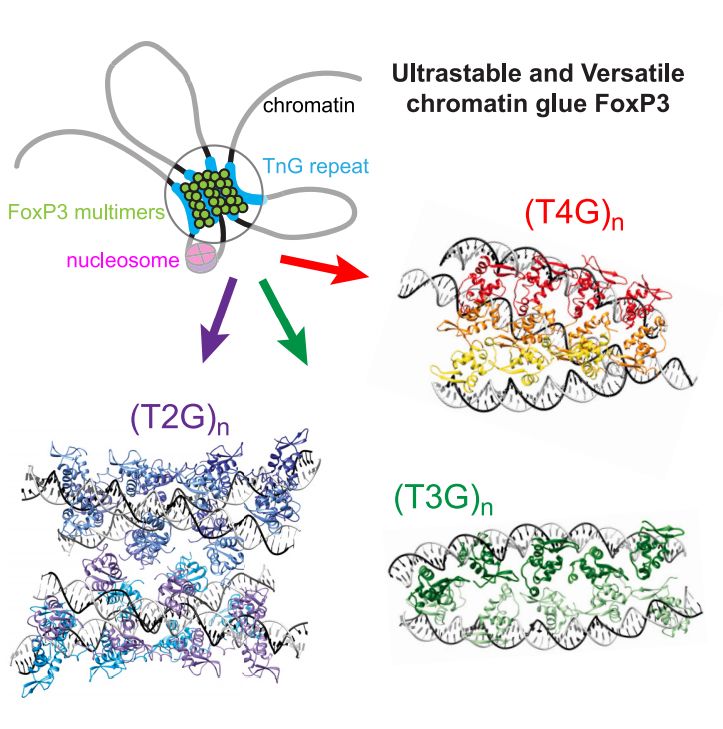
▶️FoxP3 recognizes a variety of TnG repeat microsatellites
▶️FoxP3 multimers are ultrastable, can bridge 2–4 DNA duplexes
▶️Nucleosomes can facilitate FoxP3 assembly by inducing local DNA bending






