@SEMC_NYSBC. Co-founder and CEO of http://OpenProtein.AI. Opinions are my own.
PoET-2 is also open sourced on github: github.com/OpenProteinA...
Thanks to the @openprotein.bsky.social team!

PoET-2 is also open sourced on github: github.com/OpenProteinA...
Thanks to the @openprotein.bsky.social team!
www.openprotein.ai/early-access...
www.openprotein.ai/early-access...
Sign up now: www.openprotein.ai/early-access...
Sign up now: www.openprotein.ai/early-access...
We’re solving this at OpenProtein.AI. Check out our upcoming indel design tool! 🤩 1/4
@openprotein.bsky.social
We’re solving this at OpenProtein.AI. Check out our upcoming indel design tool! 🤩 1/4
@openprotein.bsky.social
🔗 docs.openprotein.ai/walkthroughs...
Sign up for OpenProtein.AI: www.openprotein.ai/early-access...
🔗 docs.openprotein.ai/walkthroughs...
Sign up for OpenProtein.AI: www.openprotein.ai/early-access...
Check out our new walkthrough on inverse folding: docs.openprotein.ai/walkthroughs...
Check out our new walkthrough on inverse folding: docs.openprotein.ai/walkthroughs...
PoET-2 is changing the game in computational protein design, slashing experimental data needs by 30x! 🚀
learn more: www.synbiobeta.com/read/protein...
#ProteinDesign #BiotechInnovation #AIRevolution

PoET-2 is changing the game in computational protein design, slashing experimental data needs by 30x! 🚀
learn more: www.synbiobeta.com/read/protein...
#ProteinDesign #BiotechInnovation #AIRevolution
In new work led by @liambai.bsky.social and me, we explore how sparse autoencoders can help us understand biology—going from mechanistic interpretability to mechanistic biology.
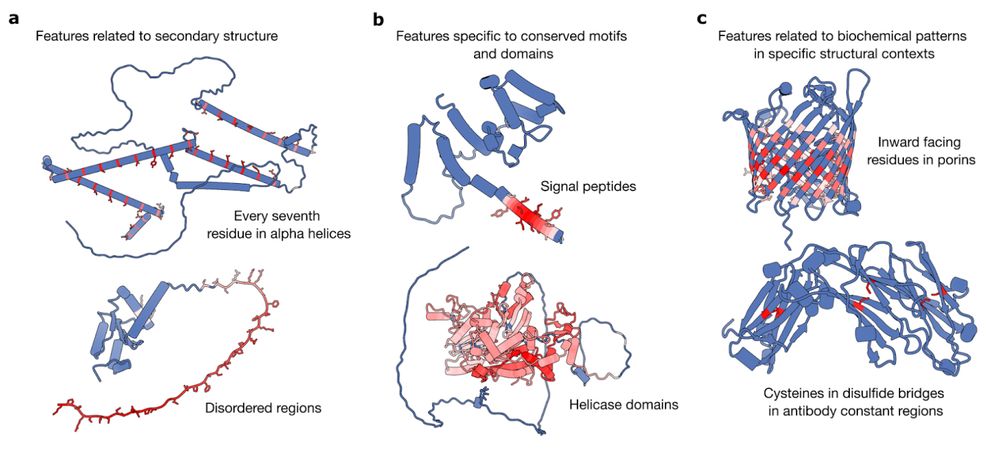
In new work led by @liambai.bsky.social and me, we explore how sparse autoencoders can help us understand biology—going from mechanistic interpretability to mechanistic biology.
Our preprint: www.biorxiv.org/content/10.1...
Visualizer: interprot.com
Github: github.com/etowahadams/...
HuggingFace: huggingface.co/liambai/Inte...

Our preprint: www.biorxiv.org/content/10.1...
Visualizer: interprot.com
Github: github.com/etowahadams/...
HuggingFace: huggingface.co/liambai/Inte...
@biorxivpreprint.bsky.social !
Thanks to hard work by Robert Kiewisz and our many collaborators!
www.biorxiv.org/content/10.1...

@biorxivpreprint.bsky.social !
Thanks to hard work by Robert Kiewisz and our many collaborators!
www.biorxiv.org/content/10.1...
If you can't make it, I'll also be presenting at the Berger Lab seminar @mitofficial.bsky.social on Wednesday!
If you can't make it, I'll also be presenting at the Berger Lab seminar @mitofficial.bsky.social on Wednesday!


