He/His/Him. Almost cartainly not D.B. Cooper
ShechnerLab.org
I had an absolute blast at the Fusion “Genome Regulation Through RNA” meeting back in 2024, and psyched to see its upcoming return! Let’s geek out about all things Chromatin and RNA together, in Cancun!
Talk abstract deadline: 10/17
Details👇


I had an absolute blast at the Fusion “Genome Regulation Through RNA” meeting back in 2024, and psyched to see its upcoming return! Let’s geek out about all things Chromatin and RNA together, in Cancun!
Talk abstract deadline: 10/17
Details👇

tinyurl.com/ydn6e3ac
tinyurl.com/ydn6e3ac

















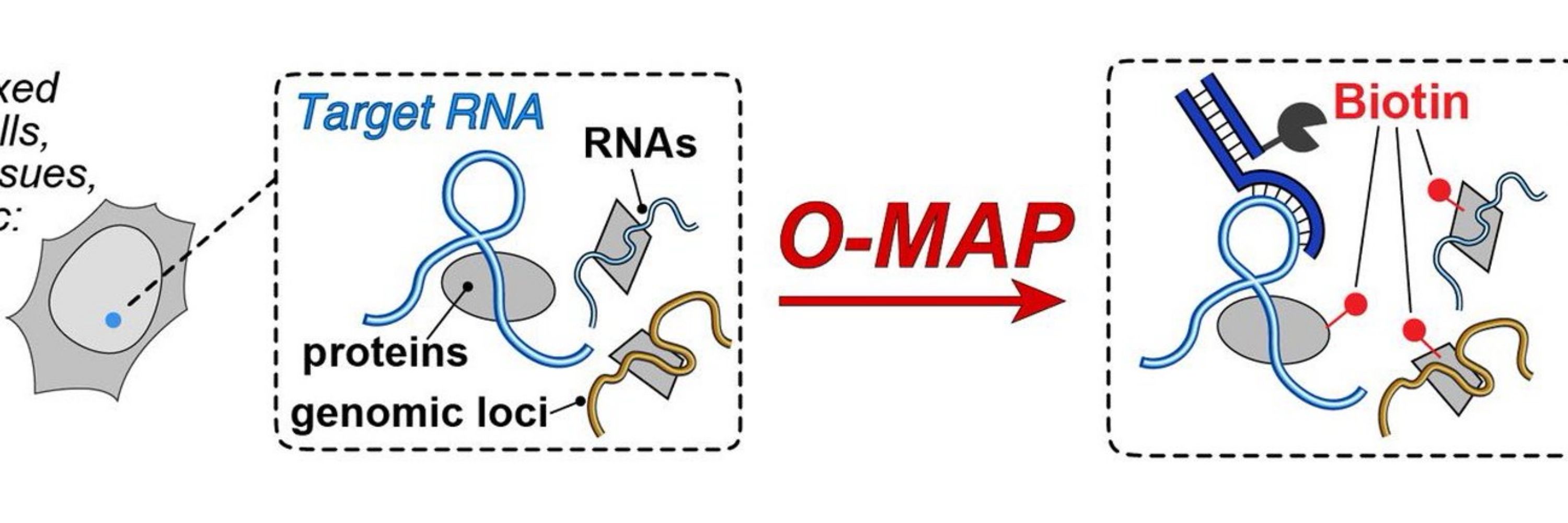
tinyurl.com/yurzfyjp

tinyurl.com/yurzfyjp
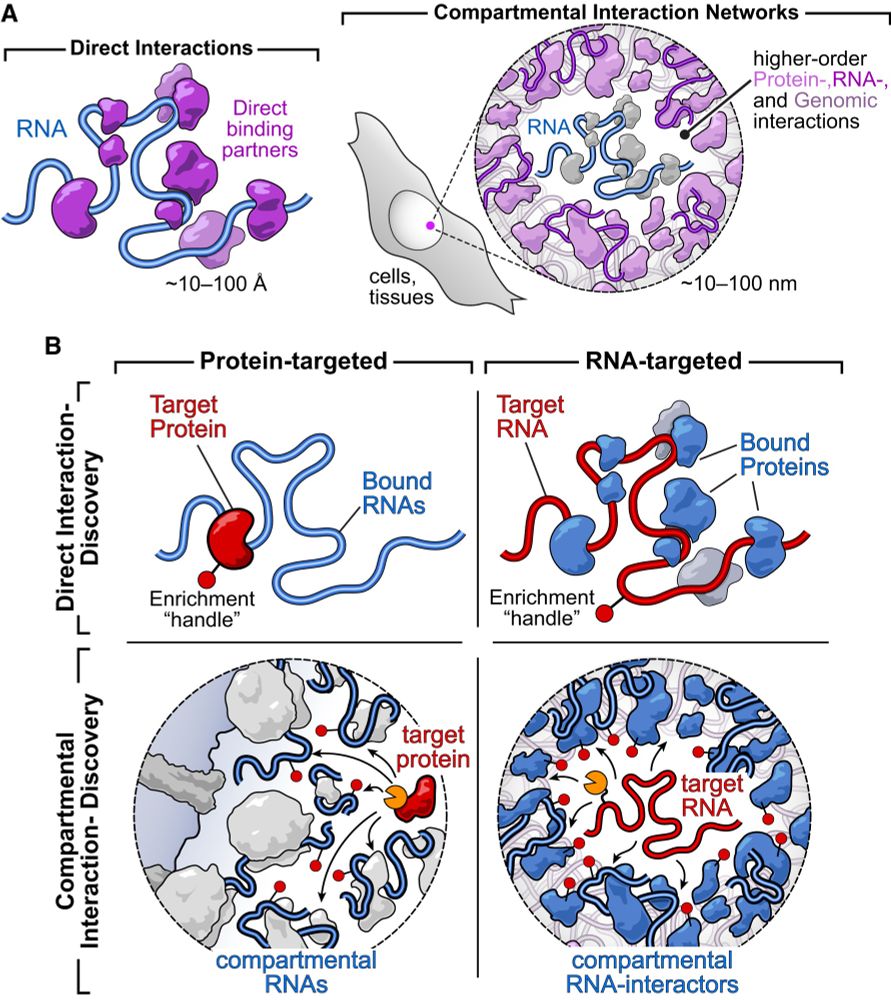
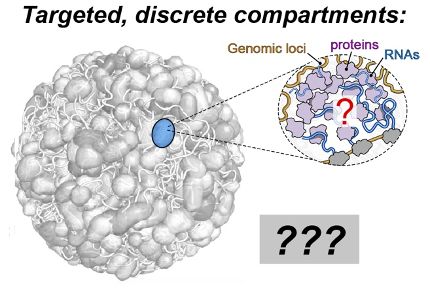
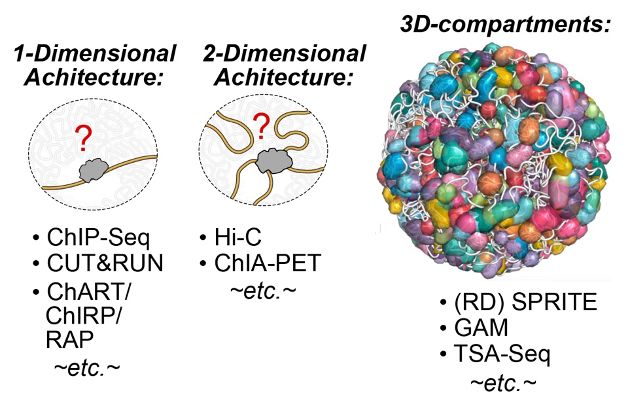
If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)
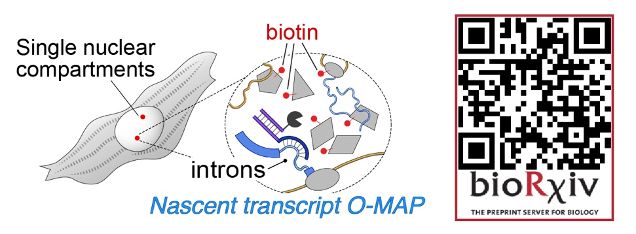
If you've ever wanted to dissect the subnuclear "neighborhood" around an individual locus, read on! (1/30)


