(meta)Genomics, bioinformatics
Overall we do not know why this is happening, but might highlight a lineage that is better adapted to gut conditions later in life (2/2)
Overall we do not know why this is happening, but might highlight a lineage that is better adapted to gut conditions later in life (2/2)
Ruminoccocus gravus is a common member of the gut microbiome. When we looked into the genetics of this bacterium, we saw that older individuals harbored bacteria of similar genetics makeup, while this was not observed in younger people. (1/2)
Ruminoccocus gravus is a common member of the gut microbiome. When we looked into the genetics of this bacterium, we saw that older individuals harbored bacteria of similar genetics makeup, while this was not observed in younger people. (1/2)
We provide a list of strain-phenotype associations and large phylogenies, hoping they’ll spark further interest in the community. (7/n)
We provide a list of strain-phenotype associations and large phylogenies, hoping they’ll spark further interest in the community. (7/n)
Interestingly, individuals with prostate cancer also clustered here! (6/n)

Interestingly, individuals with prostate cancer also clustered here! (6/n)
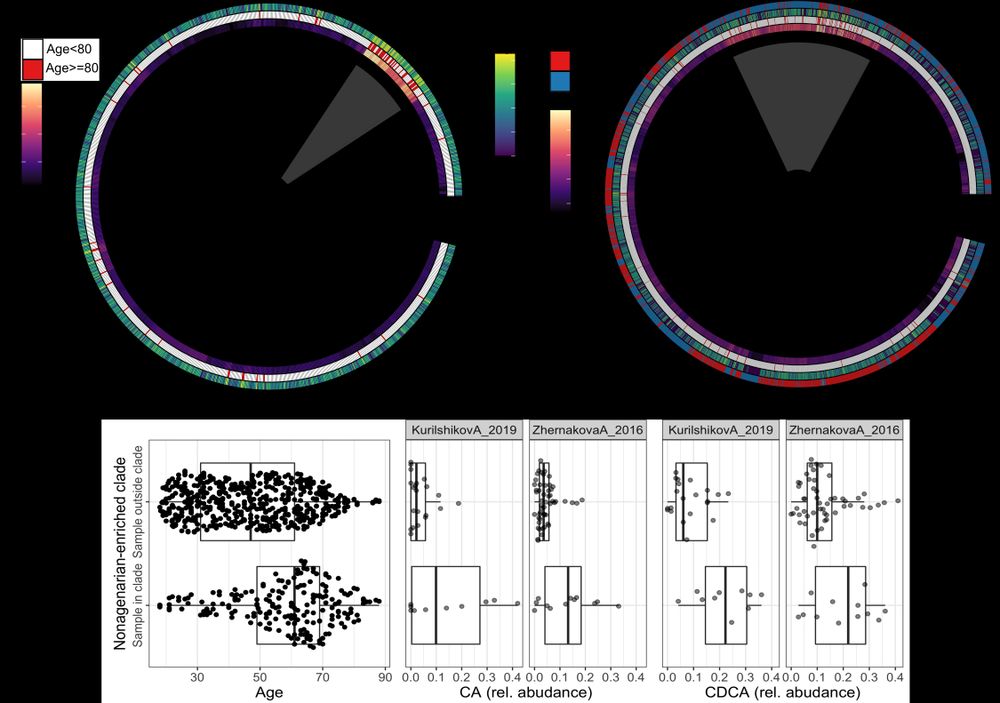
Association analysis revealed multiple phenotype–species links, including strong, reproducible associations between R. gnavus and age, and C. aerofaciens and melanoma. (4/n)
Association analysis revealed multiple phenotype–species links, including strong, reproducible associations between R. gnavus and age, and C. aerofaciens and melanoma. (4/n)
Species with stronger stratification were less often shared across individuals, suggesting limited dispersal drives the observed geographic patterns. (3/n)
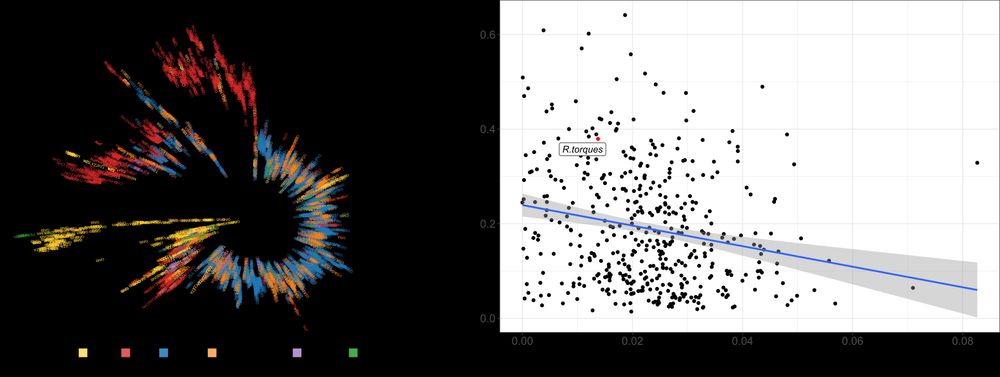
Species with stronger stratification were less often shared across individuals, suggesting limited dispersal drives the observed geographic patterns. (3/n)
We analyzed >32,000 public metagenomes, profiled species marker genes at single-nucleotide resolution with StrainPhlAn, and built ~600 species phylogenies. (2/n)
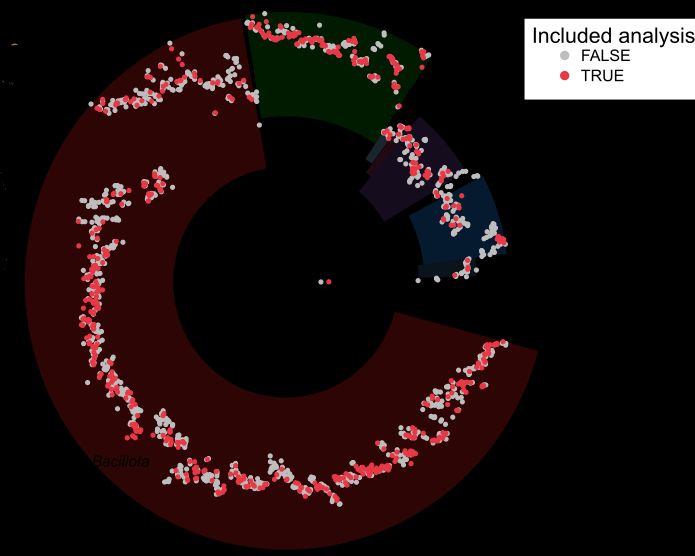
We analyzed >32,000 public metagenomes, profiled species marker genes at single-nucleotide resolution with StrainPhlAn, and built ~600 species phylogenies. (2/n)

