Member-run account of the Elizabeth Sattely Lab at Stanford University
Plant natural products | Plant-microbe interactions | Plant chemistry | Pathway discovery | Plant health | Plant-human interactions
Check out our new website and learn about the research, the people, and the publications of the Sattely Lab www.sattelylab.org

Check out our new website and learn about the research, the people, and the publications of the Sattely Lab www.sattelylab.org
We discussed the molecules responsible for the properties of medicinal plants - and how my lab studies the chemistry that plants use to make them. Check it out!
t.co/nEdpoyN8QD

We discussed the molecules responsible for the properties of medicinal plants - and how my lab studies the chemistry that plants use to make them. Check it out!
t.co/nEdpoyN8QD







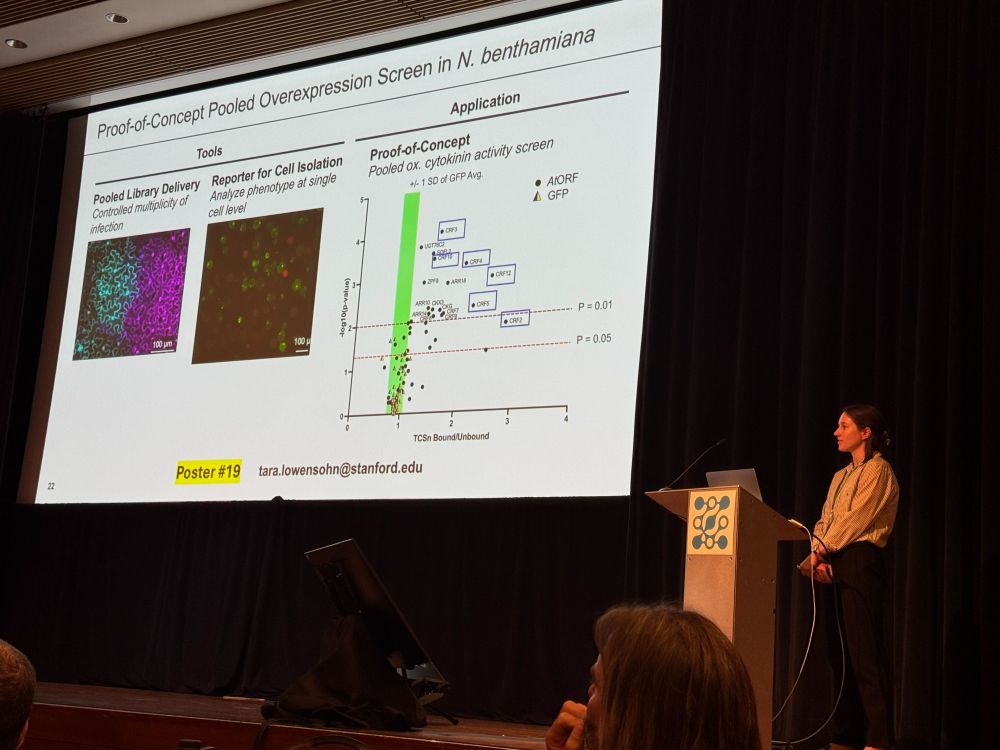
Single-gene-per-cell delivery coupled to an effective transcriptional selection system
www.biorxiv.org/content/10.1...


Fantastic team effort with the Pucker group @bpucker.bsky.social @puckerlab.bsky.social at @unibonn.bsky.social and Heretsch, Herde and Witte groups at @unihannover.bsky.social. Funded by @dfg.de
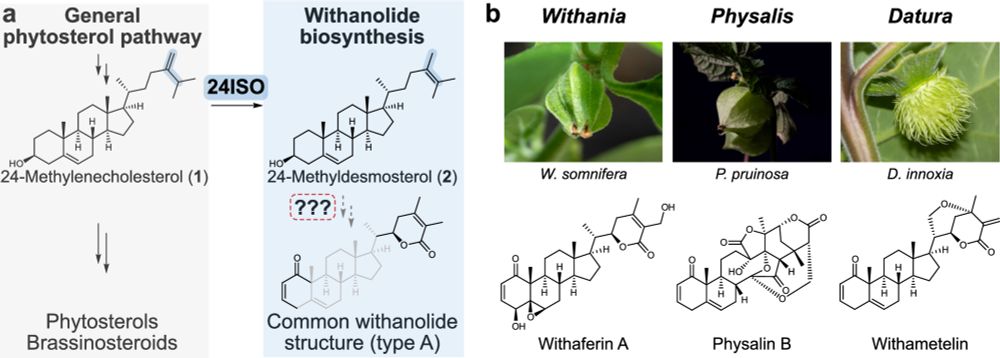
buff.ly/miccEK3
#PlantaePSRW

@sattelylab.bsky.social news.stanford.edu/stories/2025...

@sattelylab.bsky.social news.stanford.edu/stories/2025...
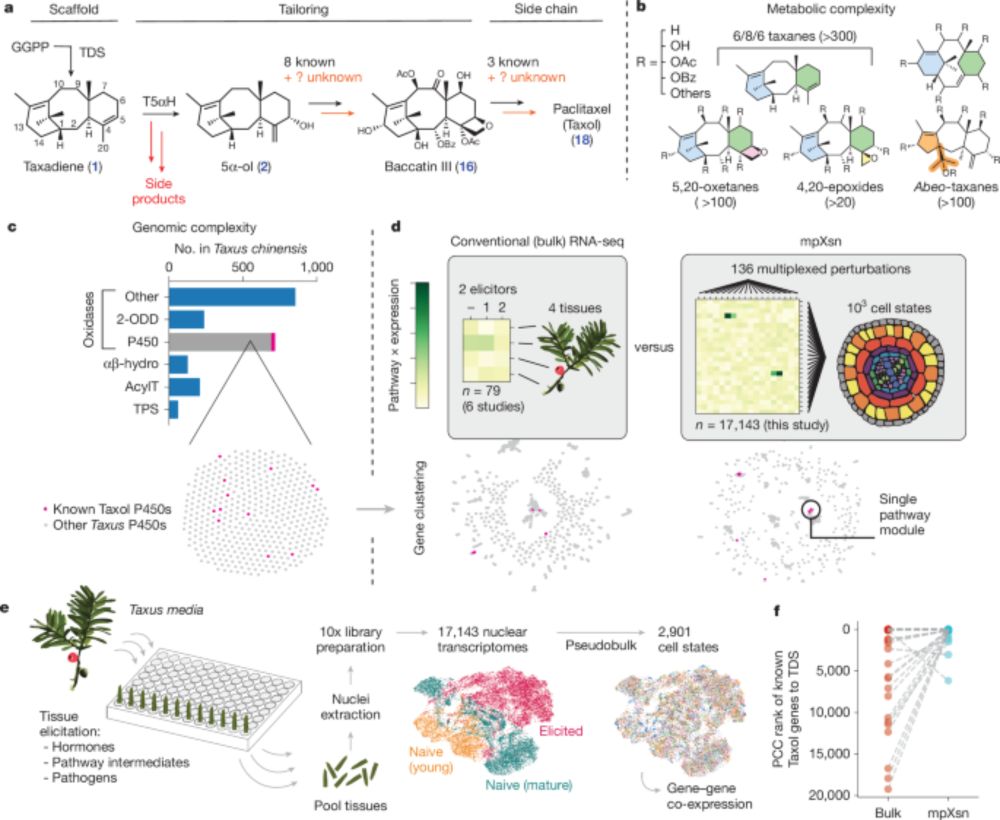
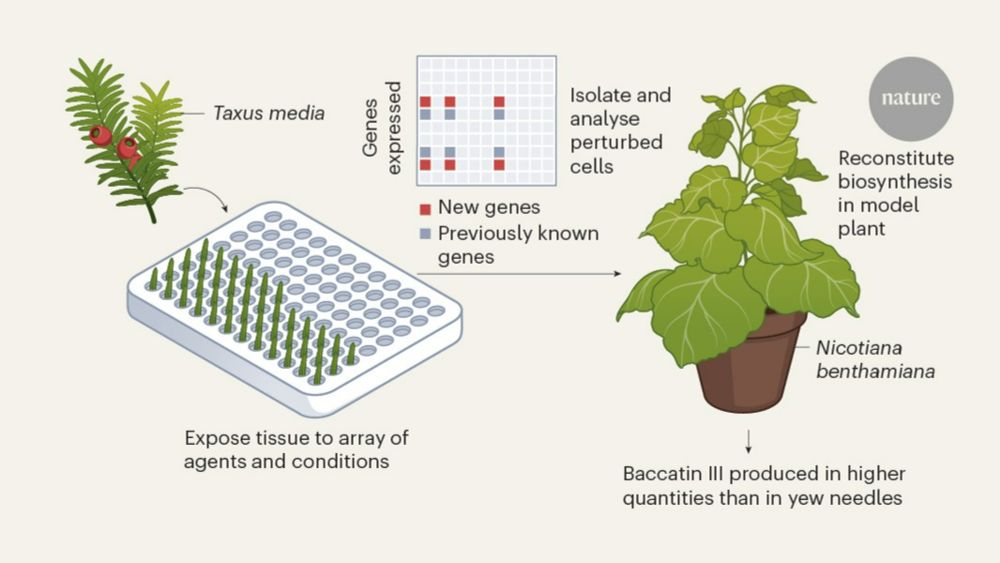
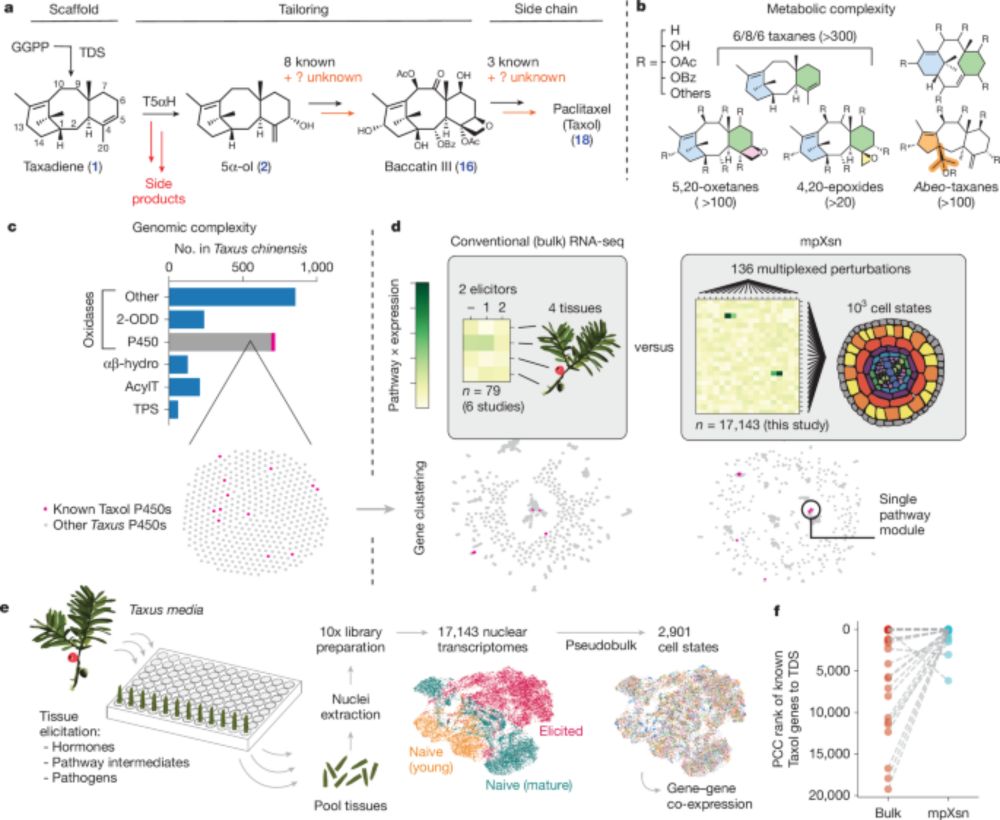
We developed a systematic strategy to activate and identify gene sets in plants. Used it to identify 8 genes in the Taxol pathway, enabling a 20-gene biosynthesis of a Taxol precursor.
Biggest surprise: a new protein that helps our oxidases produce the correct product