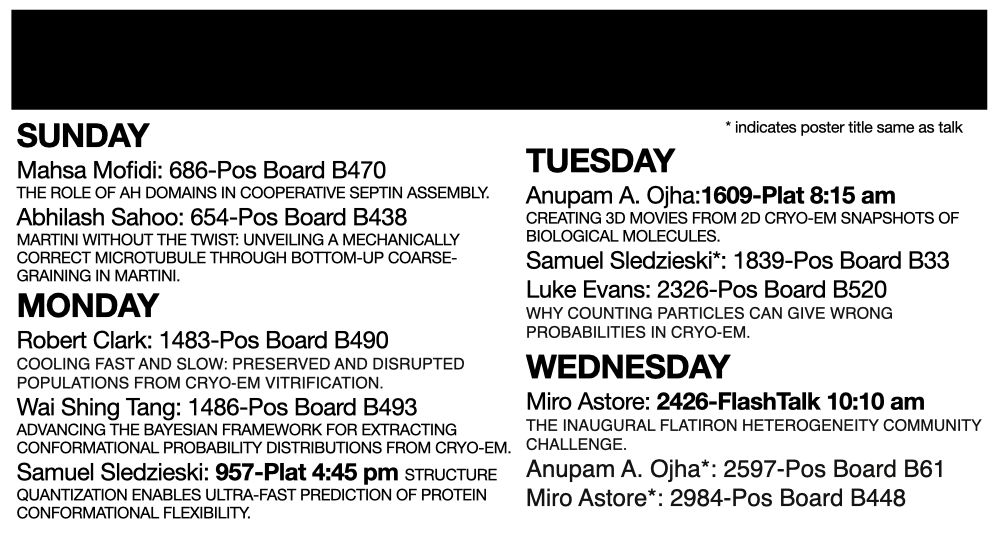
Formerly @csail.mit.edu @msftresearch.bsky.social @uconn.bsky.social
Computational systems x structure biology | he/him | https://samsl.io | 👨🏼💻

![(a) Scatter plot showing RocketSHP predicted RMSF and hetNOE, with a correlation of -0.67 and a confidence interval of [-0.68, -0.66]. (b) Line plot showing per-residue hetNOE or ReLU(1 - RMSF predicted) for the protein ACRIIA4, showing agreement between the two. (c) Two structures of ACRIIA4, colored by hetNOE or ReLU(1 - RMSF predicted), showing agreement between the two.](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:7axvdjjgnvf66wxvnv4lyg52/bafkreidaurrdcvmpbes7hi2phcaw44pxtpf3sf6pew7hjqdhk2dmmxyia4@jpeg)

MINT predictions align with 23/24 experimentally validated oncogenic PPIs impacted by cancer mutations, and MINT estimates SARS-CoV-2 antibody cross-neutralization with high accuracy.


MINT predictions align with 23/24 experimentally validated oncogenic PPIs impacted by cancer mutations, and MINT estimates SARS-CoV-2 antibody cross-neutralization with high accuracy.
It outperforms IgBert & IgT5 in predicting antibody binding affinity and estimating antibody expression.
Fine-tuning MINT beats TITAN, PISTE and other TCR-specific models on TCR–Epitope and TCR–Epitope–MHC interaction prediction.

It outperforms IgBert & IgT5 in predicting antibody binding affinity and estimating antibody expression.
Fine-tuning MINT beats TITAN, PISTE and other TCR-specific models on TCR–Epitope and TCR–Epitope–MHC interaction prediction.
It outperforms existing PLMs in:
✅ Binary PPI classification
✅ Binding affinity prediction
✅ Mutational impact assessment
Across yeast, human, & complex PPIs, we see up to 29% gains vs. baselines! 📈

It outperforms existing PLMs in:
✅ Binary PPI classification
✅ Binding affinity prediction
✅ Mutational impact assessment
Across yeast, human, & complex PPIs, we see up to 29% gains vs. baselines! 📈
We trained MINT on 96 million high-quality PPIs (from STRING-db). Instead of masked language modeling on single sequences, we now capture interaction-specific signals.

We trained MINT on 96 million high-quality PPIs (from STRING-db). Instead of masked language modeling on single sequences, we now capture interaction-specific signals.