
Website: https://saezlab.org/
GitHub: https://github.com/saezlab/
In a cohort receiving LVADs (data from Kory Lavine’s lab), molecular changes in the map were consistent with clinical improvement, suggesting that the map may help characterize treatment responses.

In a cohort receiving LVADs (data from Kory Lavine’s lab), molecular changes in the map were consistent with clinical improvement, suggesting that the map may help characterize treatment responses.





We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape

We present an extended version of ScAPE, the method that won one of the prizes 🏆 in the @neuripsconf.bsky.social 2023 Single-Cell Perturbation Prediction challenge.
📄 preprint: doi.org/10.1101/2025...
🧬 code: github.com/scapeML/scape


📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️

📖 Preprint: www.biorxiv.org/content/10.1...
🔍 Explorer: explore.omnipathdb.org
OmniPath integrates 160+ resources for multi-omics analysis & modeling.
🧶⬇️
🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io

🔗 Manuscript: www.biorxiv.org/content/10.1...
💻 Code: partipy.readthedocs.io


- PCA + ANOVA find metabolite drivers in patient subgroups
- bioRCM spots distinct patterns; e.g., dipeptides in young vs. old kidney cancer patients
➡️ Insight into tumor microenvironment and potential biomarkers

- PCA + ANOVA find metabolite drivers in patient subgroups
- bioRCM spots distinct patterns; e.g., dipeptides in young vs. old kidney cancer patients
➡️ Insight into tumor microenvironment and potential biomarkers
- Merges intra & exo-metabolomics (= bioRCM input)
- Groups metabolites by regulation patterns (see Figure)
🧪 Applying bioRCM, we found methionine consumption and usage within the cell, which could be linked to DNA hypermethylation in kidney cancer

- Merges intra & exo-metabolomics (= bioRCM input)
- Groups metabolites by regulation patterns (see Figure)
🧪 Applying bioRCM, we found methionine consumption and usage within the cell, which could be linked to DNA hypermethylation in kidney cancer
- Filters low-quality features via CV & missingness
- Handles exometabolomics-specific normalization (e.g. growth rate, media blanks)
- Detects outliers
- Differential analysis (also for exometabolomics)
➡️ Ensures high-quality data

- Filters low-quality features via CV & missingness
- Handles exometabolomics-specific normalization (e.g. growth rate, media blanks)
- Detects outliers
- Differential analysis (also for exometabolomics)
➡️ Ensures high-quality data
🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️

🔗
www.biorxiv.org/content/10.1...
📦
saezlab.github.io/MetaProViz/
🧵 Thread ⬇️
• Unified framework for context-specific network inference
• Supports diverse omics data & network types
• Enables joint multi-sample analysis for robust, interpretable results
• Flexible: add your own constraints, hypotheses, or network models
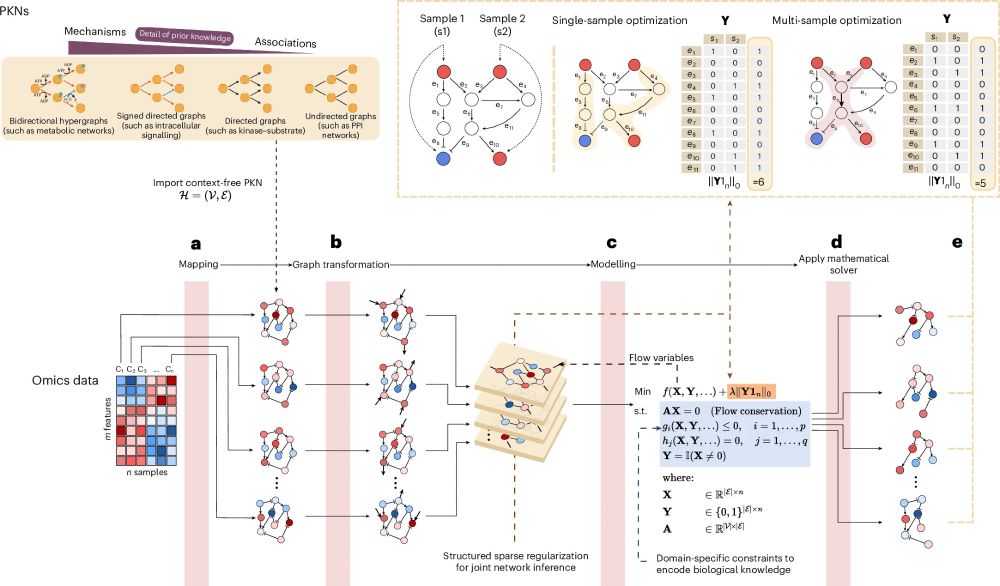
• Unified framework for context-specific network inference
• Supports diverse omics data & network types
• Enables joint multi-sample analysis for robust, interpretable results
• Flexible: add your own constraints, hypotheses, or network models
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
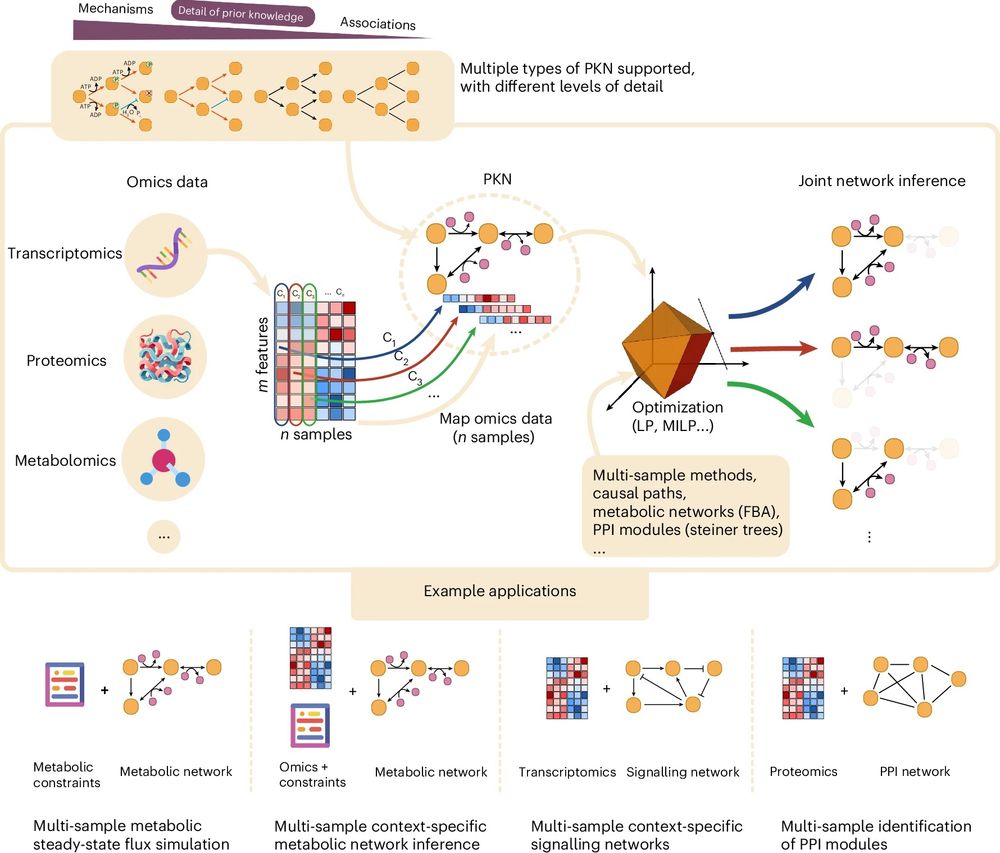
🔗 Paper: www.nature.com/articles/s42...
📖 News & Views: www.nature.com/articles/s42...
💻 Code: corneto.org
🧵 Thread 👇
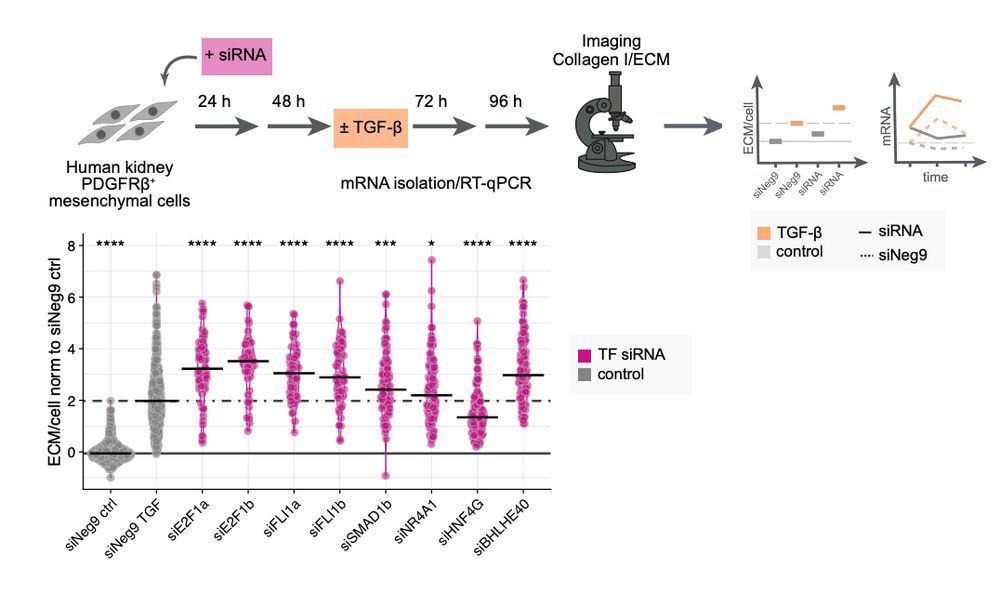
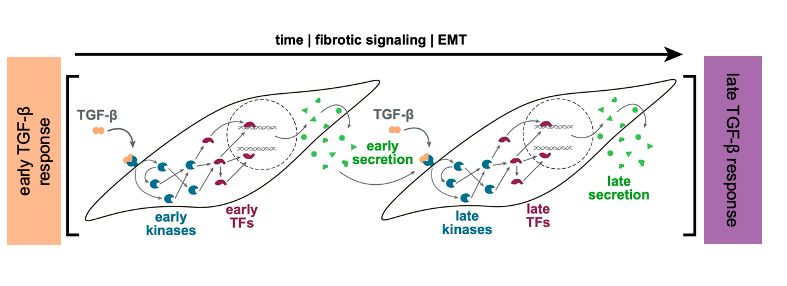
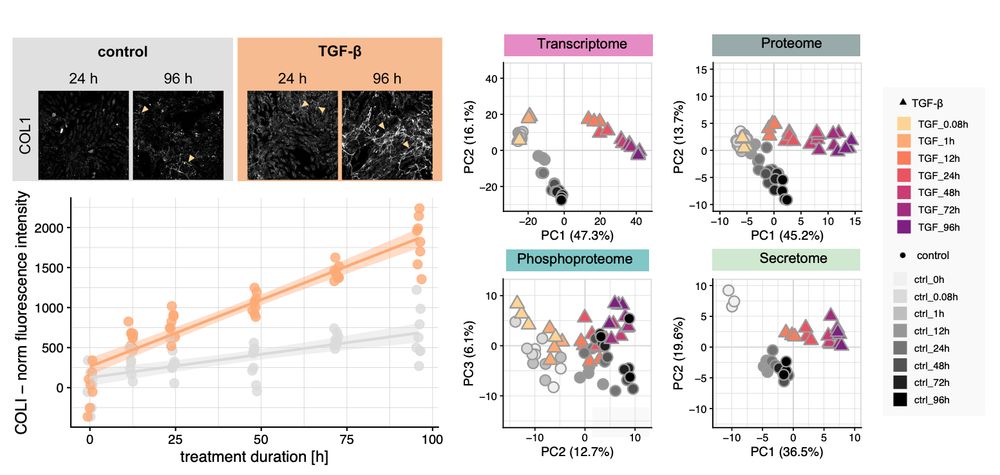
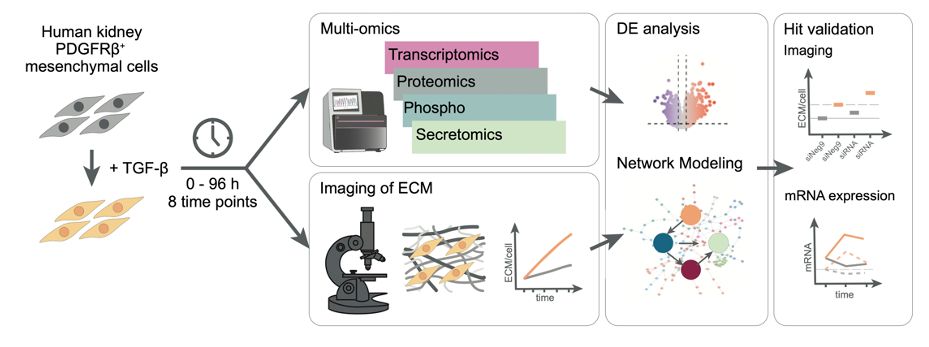

This release includes a streamlined API aligned with scverse standards, updated vignettes, and a new one on pseudotime enrichment analysis 👇
decoupler.readthedocs.io/en/latest/
This release includes a streamlined API aligned with scverse standards, updated vignettes, and a new one on pseudotime enrichment analysis 👇
decoupler.readthedocs.io/en/latest/



