

www.biorxiv.org/content/10.6...
www.biorxiv.org/content/10.6...
www.cell.com/cell/fulltex...
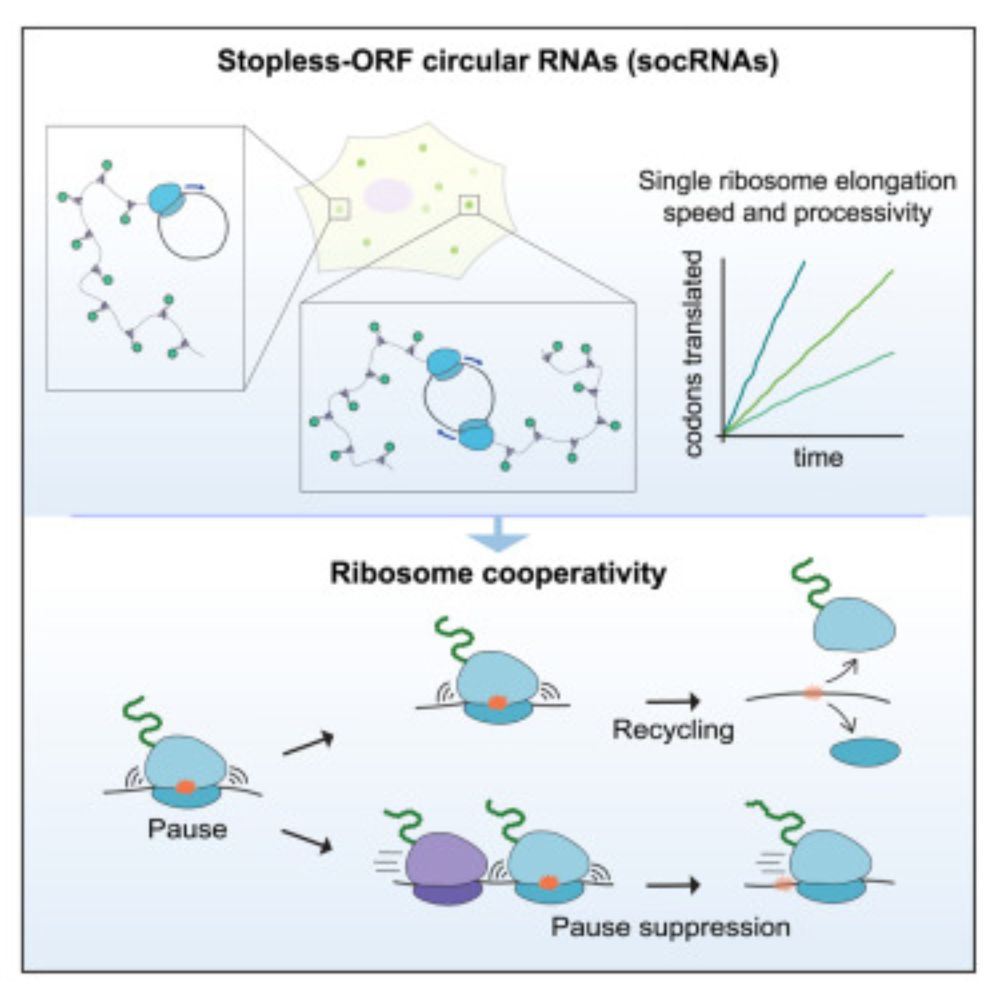
www.cell.com/cell/fulltex...
We are glad to share with you our latest publication investigating the evolution of XIST regulation in primates ES cells.
urlr.me/eyG38M
@manucazottes.bsky.social @crougeulle.bsky.social @charbel-alfeghaly.bsky.social

We are glad to share with you our latest publication investigating the evolution of XIST regulation in primates ES cells.
urlr.me/eyG38M
@manucazottes.bsky.social @crougeulle.bsky.social @charbel-alfeghaly.bsky.social
go.nature.com/49XAeGN

go.nature.com/49XAeGN
go.nature.com/4qNyxS8

go.nature.com/4qNyxS8
We uncover an unexpected role for endogenous Xist RNA in regulating X-linked genes that escape X-inactivation.

We uncover an unexpected role for endogenous Xist RNA in regulating X-linked genes that escape X-inactivation.
www.nature.com/articles/s41...

www.nature.com/articles/s41...

Huge congratulations to @huabin-zhou.bsky.social, Mike Rosen, and the brilliant @janhuemar.bsky.social @juliamaristany.bsky.social and @kieran-russell.bsky.social from our group
News: bit.ly/4avnkAr and bit.ly/3XBGVHS
Great perspective by @vram142.bsky.social +K Zhang
Huge congratulations to @huabin-zhou.bsky.social, Mike Rosen, and the brilliant @janhuemar.bsky.social @juliamaristany.bsky.social and @kieran-russell.bsky.social from our group
News: bit.ly/4avnkAr and bit.ly/3XBGVHS
Great perspective by @vram142.bsky.social +K Zhang
“𝐗𝐈𝐒𝐓 𝐃𝐫𝐢𝐯𝐞𝐬 𝐗-𝐂𝐡𝐫𝐨𝐦𝐨𝐬𝐨𝐦𝐞 𝐈𝐧𝐚𝐜𝐭𝐢𝐯𝐚𝐭𝐢𝐨𝐧 𝐚𝐧𝐝 𝐒𝐚𝐟𝐞𝐠𝐮𝐚𝐫𝐝𝐬 𝐅𝐞𝐦𝐚𝐥𝐞 𝐄𝐱𝐭𝐫𝐚𝐞𝐦𝐛𝐫𝐲𝐨𝐧𝐢𝐜 𝐂𝐞𝐥𝐥𝐬 𝐢𝐧 𝐇𝐮𝐦𝐚𝐧𝐬” 𝐢𝐬 𝐧𝐨𝐰 𝐨𝐧 𝐛𝐢𝐨𝐑𝐱𝐢𝐯!
www.biorxiv.org/content/10.1...

www.nature.com/articles/s41...

www.nature.com/articles/s41...


👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial

👉 We discover how sex and development signals are decoded at a single gene locus.
www.nature.com/articles/s41...
👇 Bluetorial
bit.ly/3VMcyNZ

bit.ly/3VMcyNZ
https://go.nature.com/3GOTQkz

https://go.nature.com/3GOTQkz
https://go.nature.com/4kfH6BC

https://go.nature.com/4kfH6BC
bit.ly/4pIA0ta
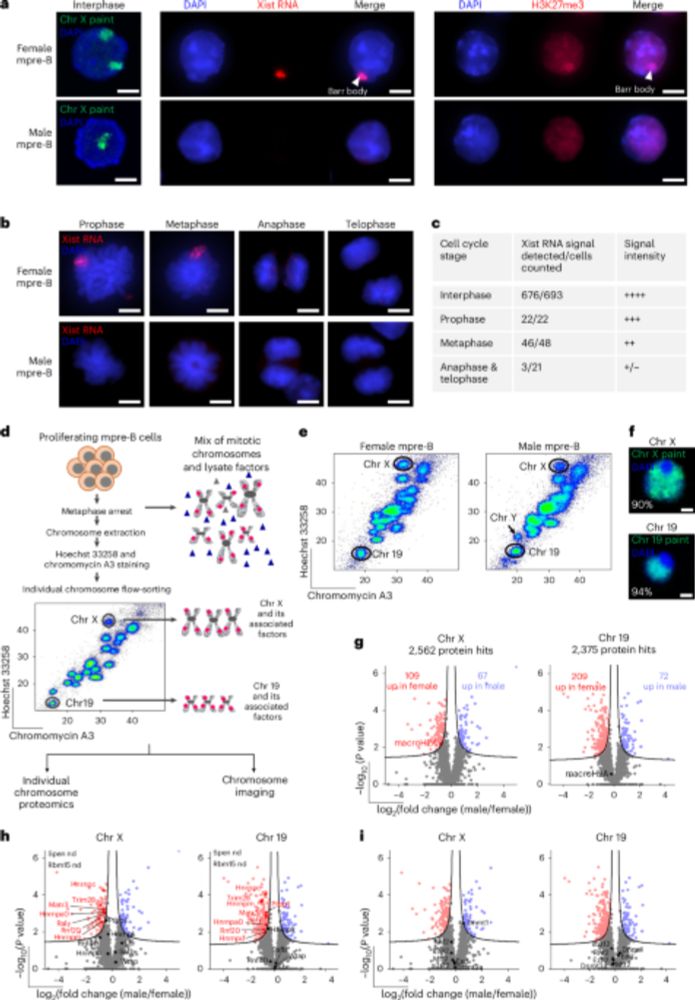
bit.ly/4pIA0ta
go.nature.com/3VmV1vp

go.nature.com/3VmV1vp

www.biorxiv.org/content/10.1...
A🧵👇
www.biorxiv.org/content/10.1...
A🧵👇


