@rgottardo.bsky.social
@markrobinsonca.bsky.social
@stephaniehicks.bsky.social
@lmweber.bsky.social
and many more.
@rgottardo.bsky.social
@markrobinsonca.bsky.social
@stephaniehicks.bsky.social
@lmweber.bsky.social
and many more.
@unil.bsky.social #chuv @owkin.bsky.social
@unil.bsky.social #chuv @owkin.bsky.social
🔗 www.nature.com/articles/s41...
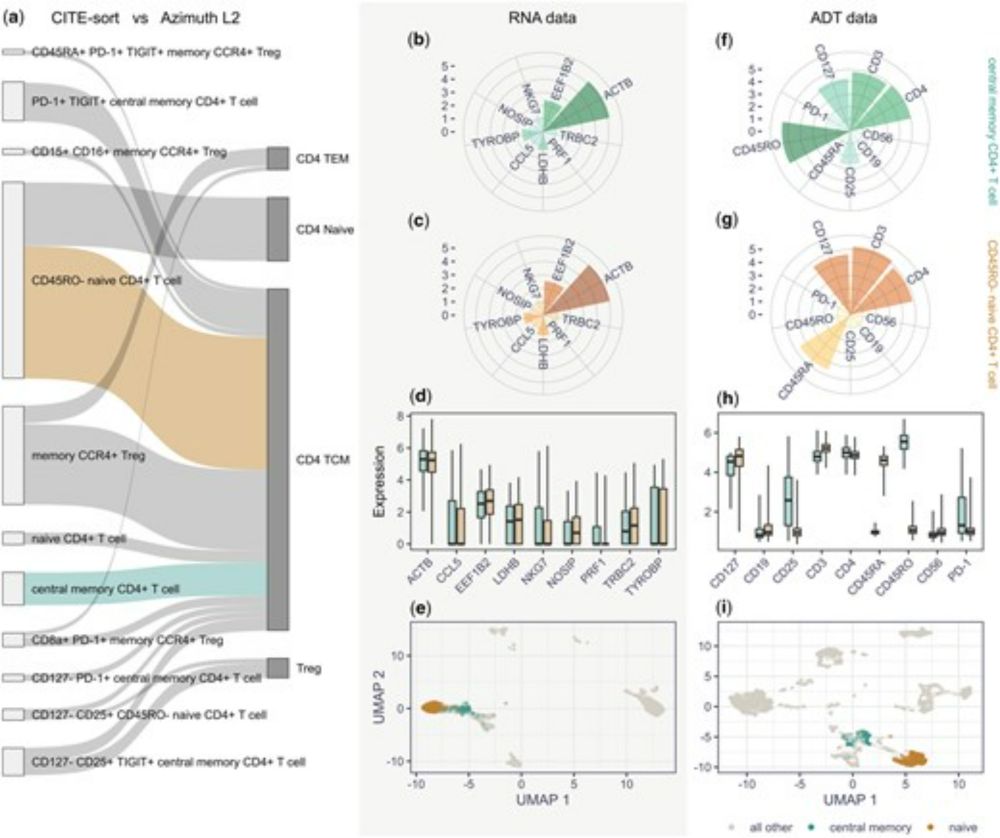
Frustrated by noisy CITE-seq data?
ADTNorm helps normalize it and introduces a staining index to assess antibody quality.
Great work by former postdoc Ye Zheng. Supported by @hipcproject.bsky.social
#CITEseq #SingleCell #ADTNorm

🔗 www.nature.com/articles/s41...
Frustrated by noisy CITE-seq data?
ADTNorm helps normalize it and introduces a staining index to assess antibody quality.
Great work by former postdoc Ye Zheng. Supported by @hipcproject.bsky.social
#CITEseq #SingleCell #ADTNorm
🧠📍 Benchmarking clustering in spatial transcriptomics isn’t easy — but here is SACCELERATOR
Preprint: www.biorxiv.org/content/10.1...
With markrobinsonca.bsky.social, Naveed Ishaque, Ahmed Mahfouz, Brian Long

🧠📍 Benchmarking clustering in spatial transcriptomics isn’t easy — but here is SACCELERATOR
Preprint: www.biorxiv.org/content/10.1...
With markrobinsonca.bsky.social, Naveed Ishaque, Ahmed Mahfouz, Brian Long
www.biorxiv.org/content/10.1...
Collaboration with @khomicsko.bsky.social
cc: @10xgenomics.bsky.social
#SpatialTranscriptomics
www.biorxiv.org/content/10.1...
Collaboration with @khomicsko.bsky.social
cc: @10xgenomics.bsky.social
#SpatialTranscriptomics
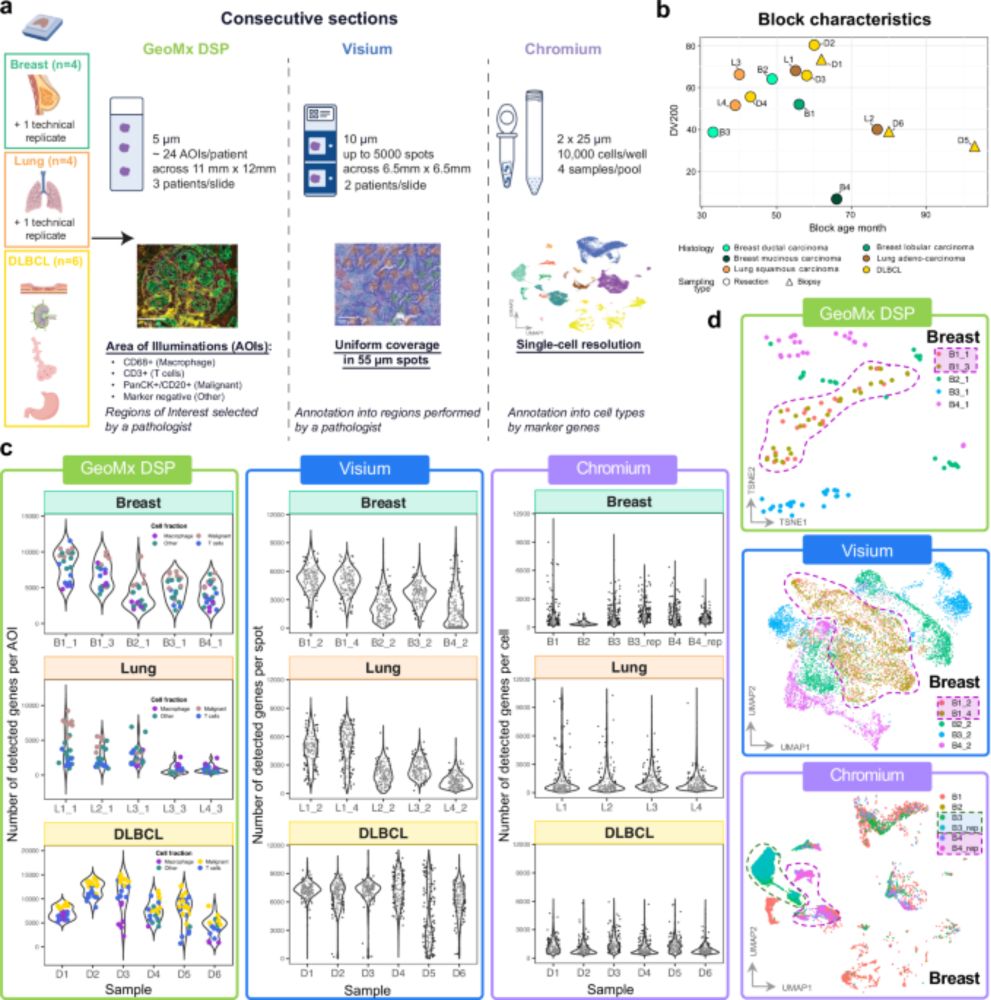
📄 www.biorxiv.org/content/10.1...
#CancerResearch #SpatialTranscriptomics #SingleCell #PrecisionOncology #AI
cc: @owkin.bsky.social @10xgenomics.bsky.social
📄 www.biorxiv.org/content/10.1...
#CancerResearch #SpatialTranscriptomics #SingleCell #PrecisionOncology #AI
cc: @owkin.bsky.social @10xgenomics.bsky.social
📄 www.nature.com/articles/s41...
#SpatialTranscriptomics #CancerResearch cc: @owkin.bsky.social @10xgenomics.bsky.social
📄 www.nature.com/articles/s41...
#SpatialTranscriptomics #CancerResearch cc: @owkin.bsky.social @10xgenomics.bsky.social
news.yale.edu/2025/03/27/e...

#bioinformatics
academic.oup.com/bioinformati...
#bioinformatics
academic.oup.com/bioinformati...
go.bsky.app/AozZWPx
go.bsky.app/AozZWPx
www.chuv.ch/fr/chuv-home... Paper on @naturecomms.bsky.social www.nature.com/articles/s41...
www.chuv.ch/fr/chuv-home... Paper on @naturecomms.bsky.social www.nature.com/articles/s41...

www.ucsf.edu/news/2024/11...

www.ucsf.edu/news/2024/11...
go.bsky.app/73rcuJn
go.bsky.app/73rcuJn
actu.epfl.ch/news/two-app...

actu.epfl.ch/news/two-app...
BUT why didn't they cite previous work in this space by Raftery and colleagues: www.science.org/doi/full/10....
journals.ametsoc.org/view/journal...
and many other papers!
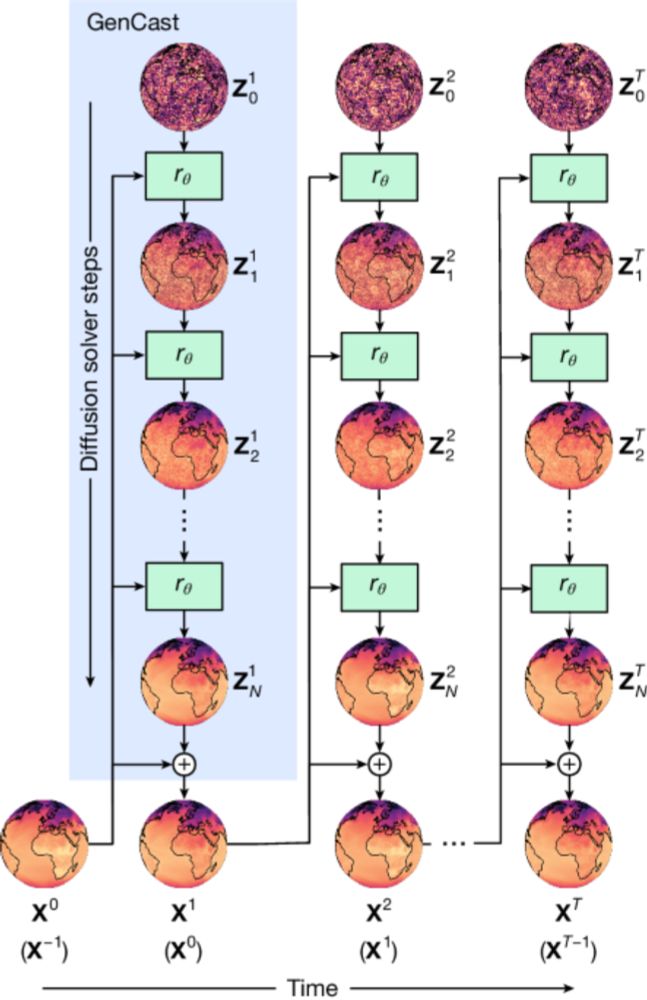
BUT why didn't they cite previous work in this space by Raftery and colleagues: www.science.org/doi/full/10....
journals.ametsoc.org/view/journal...
and many other papers!
go.bsky.app/AozZWPx
go.bsky.app/AozZWPx
go.bsky.app/73rcuJn
Reach out if you want to be added!
#AI #ML #DataScience #Omics #SingleCell #SpatialBiology
go.bsky.app/73rcuJn
Please reach out if you want to be added! The more the better.
#AI #ML #DataScience #Omics #SingleCell #SpatialBiology
go.bsky.app/73rcuJn
go.bsky.app/73rcuJn
Reach out if you want to be added!
#AI #ML #DataScience #Omics #SingleCell #SpatialBiology
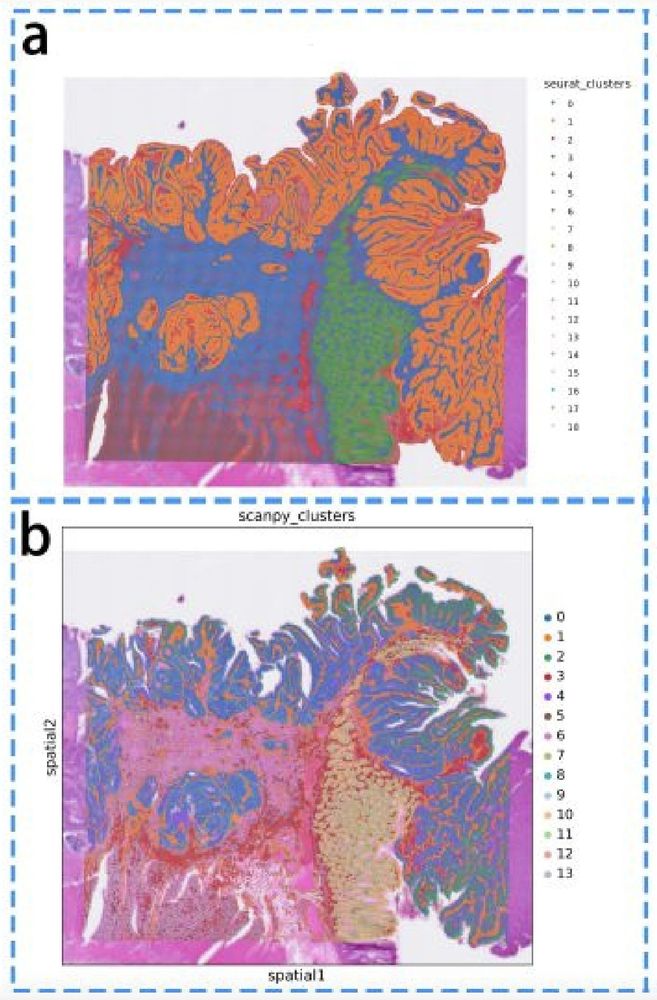
13 vs. 20 clusters. And qualitatively different results, corroborating what we saw in biorxiv.org/content/10.1...
go.bsky.app/73rcuJn
Please reach out if you want to be added! The more the better.
#AI #ML #DataScience #Omics #SingleCell #SpatialBiology
go.bsky.app/73rcuJn
go.bsky.app/73rcuJn
Please reach out if you want to be added! The more the better.
#AI #ML #DataScience #Omics #SingleCell #SpatialBiology
go.bsky.app/73rcuJn
go.bsky.app/73rcuJn