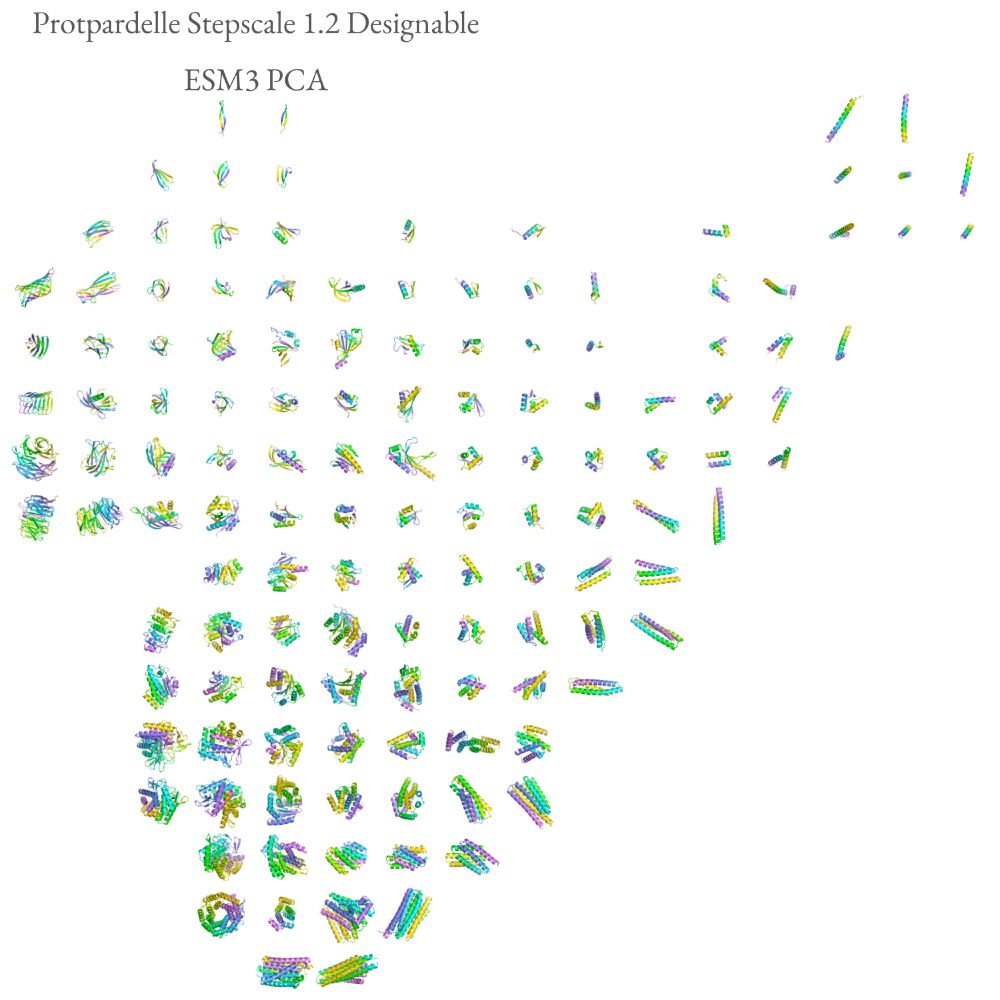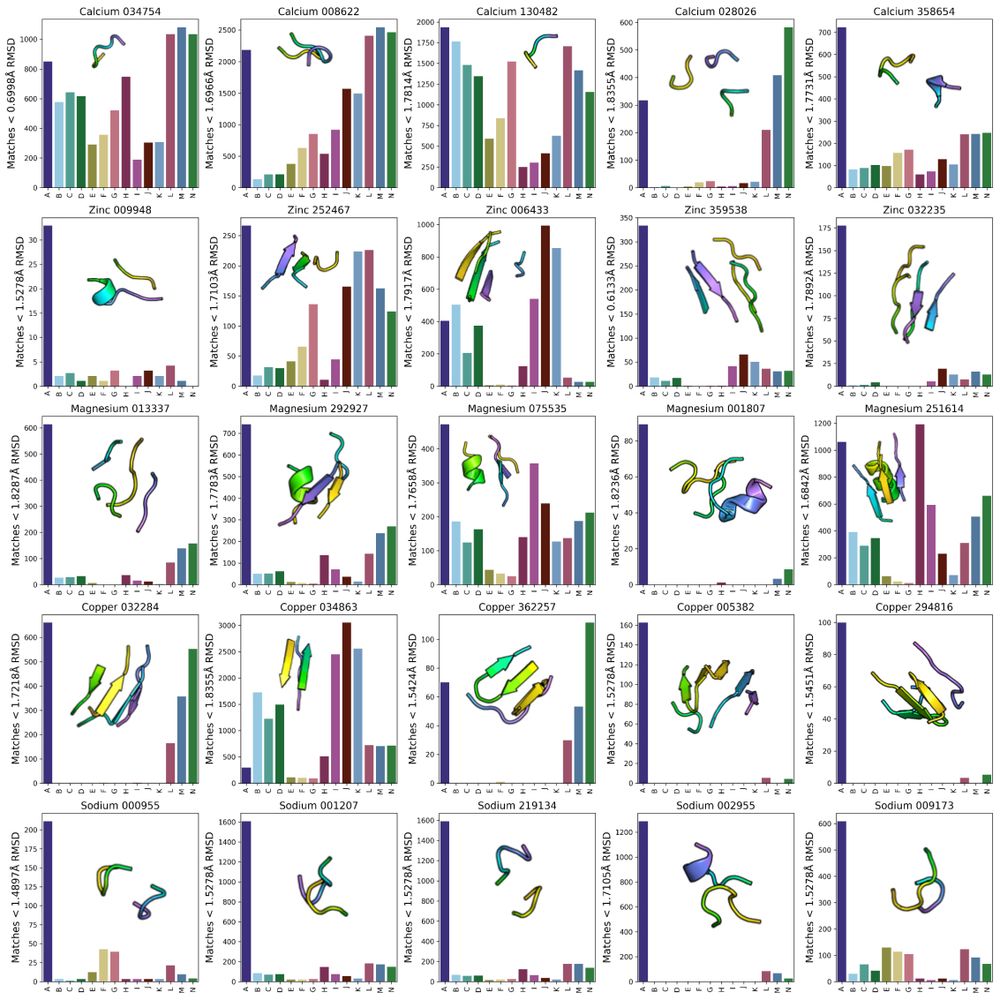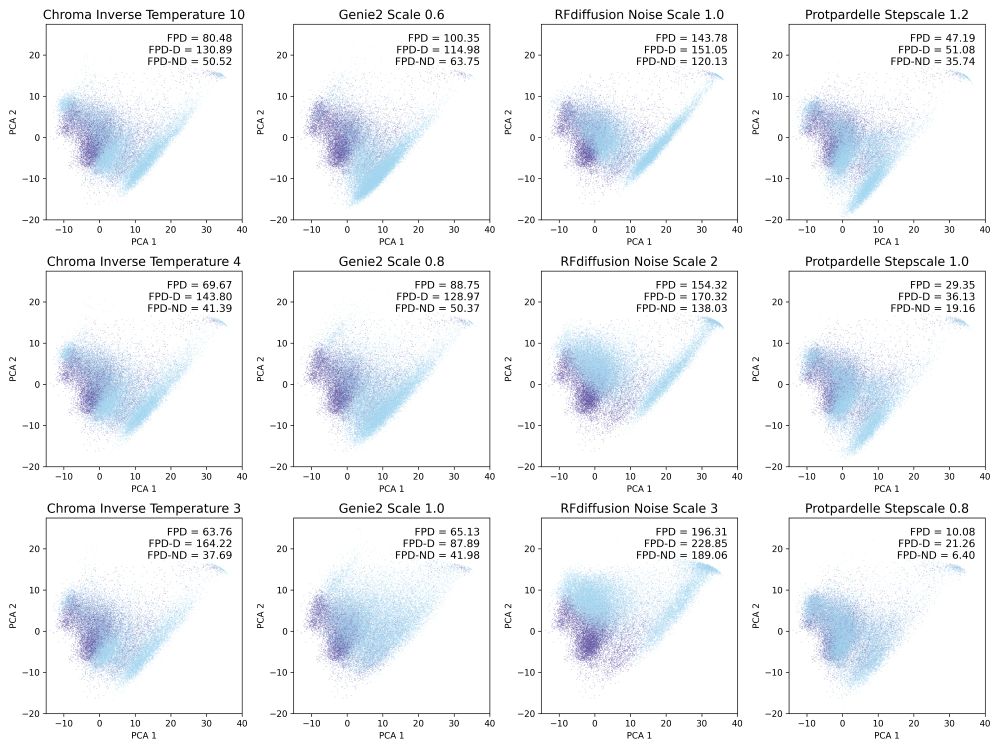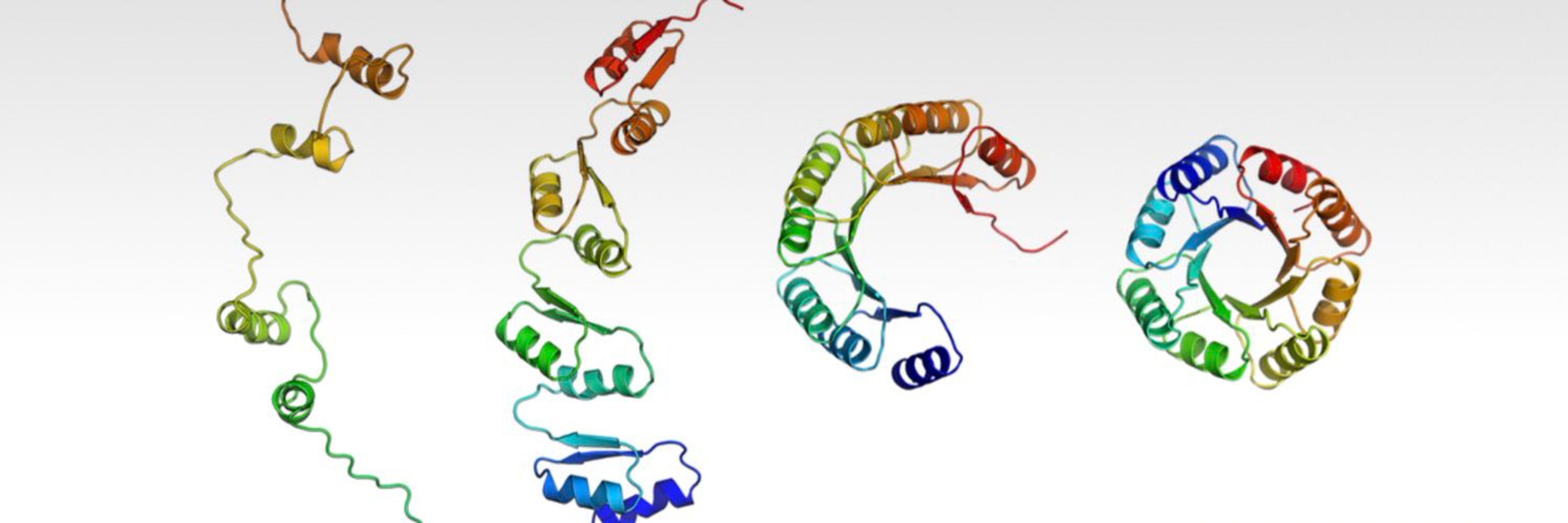




Preprint: www.biorxiv.org/content/10.1...
Have fun sampling and training!

Preprint: www.biorxiv.org/content/10.1...
Have fun sampling and training!


Code: github.com/ProteinDesig...
Dataset: zenodo.org/records/1458...
Code: github.com/ProteinDesig...
Dataset: zenodo.org/records/1458...