
Before: Moscow State University Neuroscience'20 :: Max Planck Florida Institute of Neuroscience'23 :: MS from Scheiffele lab
Phase contrast imaging started at DIV1. One frame per 20 min.
Phase contrast imaging started at DIV1. One frame per 20 min.
@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...

@aakaran31.bsky.social and @rivaselenarivas.bsky.social
link.springer.com/article/10.1...
www.cell.com/cell-reports...

www.cell.com/cell-reports...

To predict the behaviour of a primate, would you rather base your guess on a closely related species or one with a similar brain shape? We looked at brains & behaviours of 70 species, you’ll be surprised!
🧵Thread on our new preprint with @r3rt0.bsky.social , doi.org/10.1101/2025...
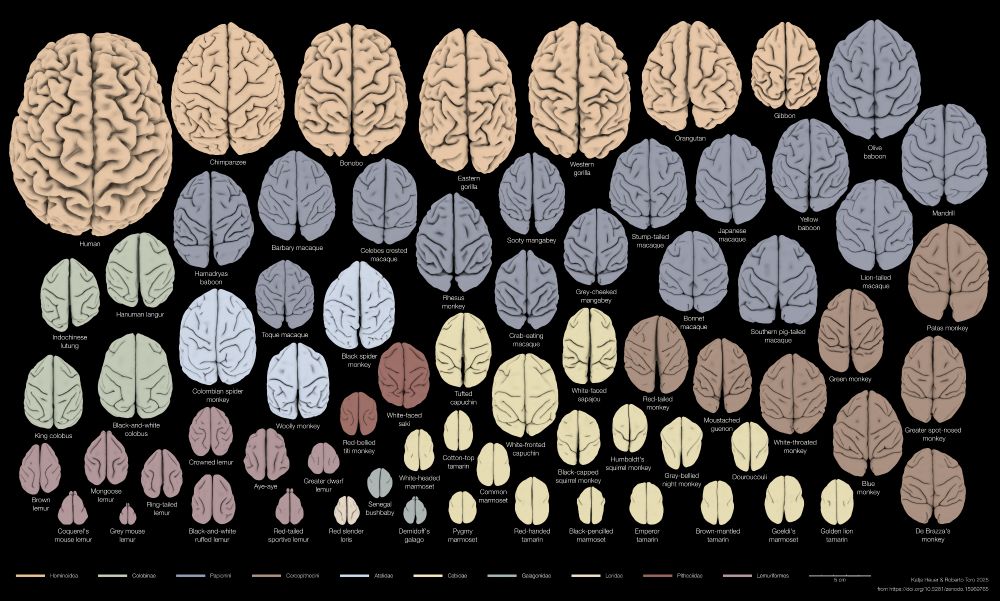
To predict the behaviour of a primate, would you rather base your guess on a closely related species or one with a similar brain shape? We looked at brains & behaviours of 70 species, you’ll be surprised!
🧵Thread on our new preprint with @r3rt0.bsky.social , doi.org/10.1101/2025...
Tooting my own horn here, but if you’re interested in some of the practical considerations mentioned at the end of the article, you can check out this perspective from earlier in my postdoc: www.cell.com/neuron/fullt...
Tooting my own horn here, but if you’re interested in some of the practical considerations mentioned at the end of the article, you can check out this perspective from earlier in my postdoc: www.cell.com/neuron/fullt...
10 yrs of mouse social networks + 1.25 yrs of acoustic data ➡️ insight into vocalization & sociality in a wild population of your favorite lab model 🐁
paper: bit.ly/4n93yyD
data: bit.ly/4lfFBEk
code: bit.ly/4kNnMwx
#bioacoustics #neuroskyence
1/8

10 yrs of mouse social networks + 1.25 yrs of acoustic data ➡️ insight into vocalization & sociality in a wild population of your favorite lab model 🐁
paper: bit.ly/4n93yyD
data: bit.ly/4lfFBEk
code: bit.ly/4kNnMwx
#bioacoustics #neuroskyence
1/8
Under the microscope, this fish gill structure looks just like a sea pen.
Fascinating how nature keeps circling back to the same shapes.
Want to learn how gills develop?
👉 www.biorxiv.org/content/10.1...
#FluorescenceFriday #Zebrafish


Under the microscope, this fish gill structure looks just like a sea pen.
Fascinating how nature keeps circling back to the same shapes.
Want to learn how gills develop?
👉 www.biorxiv.org/content/10.1...
#FluorescenceFriday #Zebrafish
Keywords: patch clamp ephys, opto, mouse behavior, (in vivo) voltage imaging. Would love to return to the Basal Ganglia.
Sharing appreciated, and happy #FluorescenceFriday !
Keywords: patch clamp ephys, opto, mouse behavior, (in vivo) voltage imaging. Would love to return to the Basal Ganglia.
Sharing appreciated, and happy #FluorescenceFriday !

We present iGluSnFR4f and 4s, a novel pair of genetically-encoded glutamate indicators designed for high-fidelity imaging of synaptic activity in the living brain. ⬇️
www.biorxiv.org/content/10.1...
🎥 Below: iGluSnFR4s detecting minis in cultures w/ TTX
#Neuroscience
We present iGluSnFR4f and 4s, a novel pair of genetically-encoded glutamate indicators designed for high-fidelity imaging of synaptic activity in the living brain. ⬇️
www.biorxiv.org/content/10.1...
🎥 Below: iGluSnFR4s detecting minis in cultures w/ TTX
#Neuroscience
—Synthetic, high-affinity #ChemicalBiology probes for #SuperResolution #Synapse visualization & precise mapping in neurons and brain slices—without the need for antibodies, tags, or transfection!
📢 Read more: pubs.acs.org/doi/10.1021/...
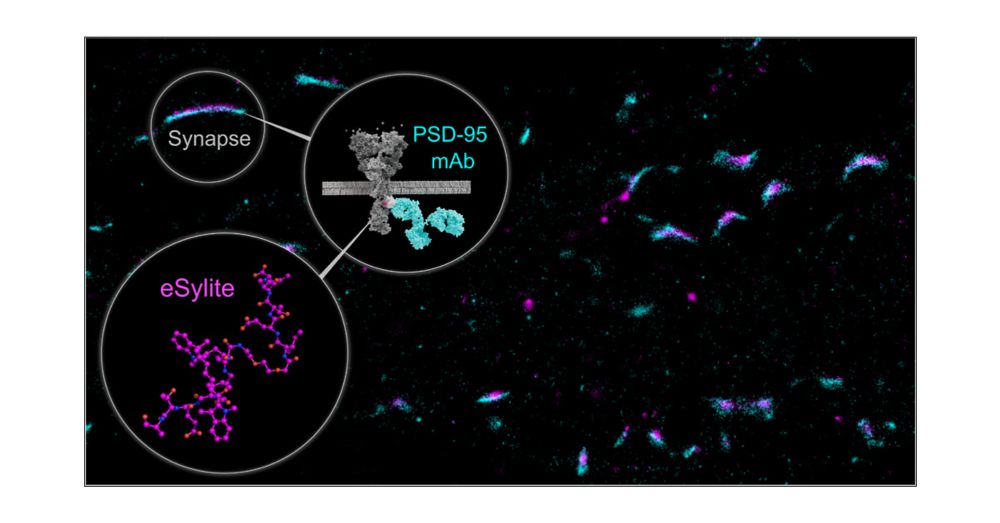
—Synthetic, high-affinity #ChemicalBiology probes for #SuperResolution #Synapse visualization & precise mapping in neurons and brain slices—without the need for antibodies, tags, or transfection!
📢 Read more: pubs.acs.org/doi/10.1021/...
🔬 Microscopists
💻 BioImage Analysts
👩🔬 Life Scientists
📅 Save the Date
The 5th edition of the Crick BioImage Analysis Symposium will take place on November 24/25th 2025.
More details to follow...
#CBIAS2025

🔬 Microscopists
💻 BioImage Analysts
👩🔬 Life Scientists
📅 Save the Date
The 5th edition of the Crick BioImage Analysis Symposium will take place on November 24/25th 2025.
More details to follow...
#CBIAS2025
#RNA #RNAsky
RNA-binding proteins and glycoRNAs form domains on the cell surface for cell-penetrating peptide entry: Cell www.cell.com/cell/fulltex...

#RNA #RNAsky
RNA-binding proteins and glycoRNAs form domains on the cell surface for cell-penetrating peptide entry: Cell www.cell.com/cell/fulltex...
Finally this work is out - we show that short sequence motifs direct proteins to specific peroxisomal sub-domains. Fantastic collaboration with @bangebalcony.bsky.social , Johannes Freitag @unimarburg.bsky.social and many more! @uio.no @cancelluio.bsky.social

Finally this work is out - we show that short sequence motifs direct proteins to specific peroxisomal sub-domains. Fantastic collaboration with @bangebalcony.bsky.social , Johannes Freitag @unimarburg.bsky.social and many more! @uio.no @cancelluio.bsky.social
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
What would you do with this?
Now… what would you do if that also worked with 4 structures at once? 👇 #MicroSplit #preview🧵
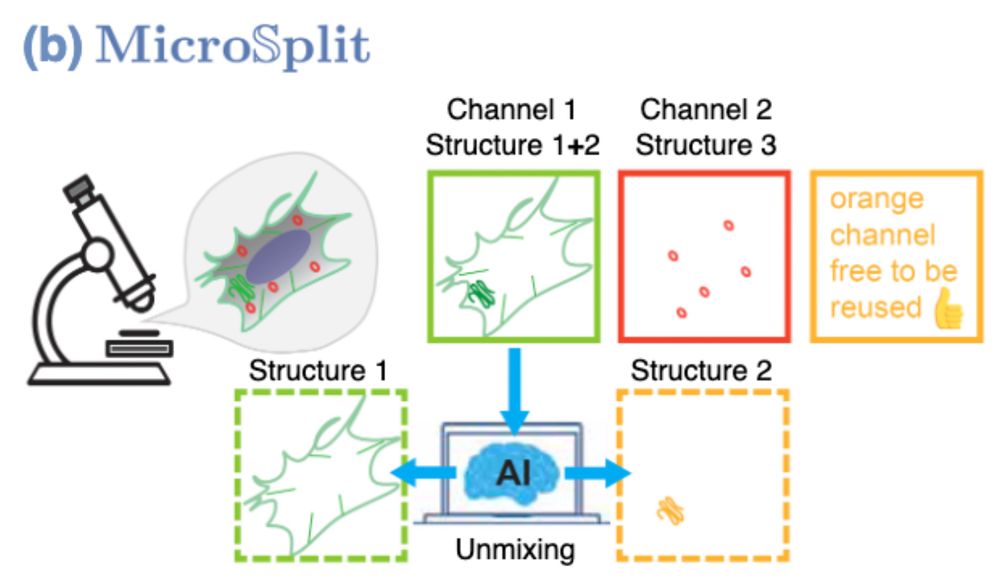
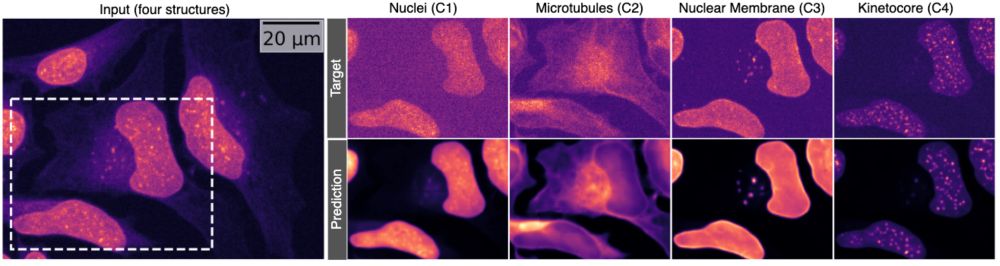
What would you do with this?
Now… what would you do if that also worked with 4 structures at once? 👇 #MicroSplit #preview🧵

My Fiji training notes are now FAIR. All online in an open format to download and use for teaching and training. All in Google Doc or PDF formats
bit.ly/4hkfEBF

My Fiji training notes are now FAIR. All online in an open format to download and use for teaching and training. All in Google Doc or PDF formats
bit.ly/4hkfEBF
🧵👇

🧵👇
Labelled with DiI, imaged with the FV3000 CLSM, & depicted using @kwolbachia.bsky.social new KTZ_bw_kawa LUT.

Labelled with DiI, imaged with the FV3000 CLSM, & depicted using @kwolbachia.bsky.social new KTZ_bw_kawa LUT.
doi.org/10.1038/s415...
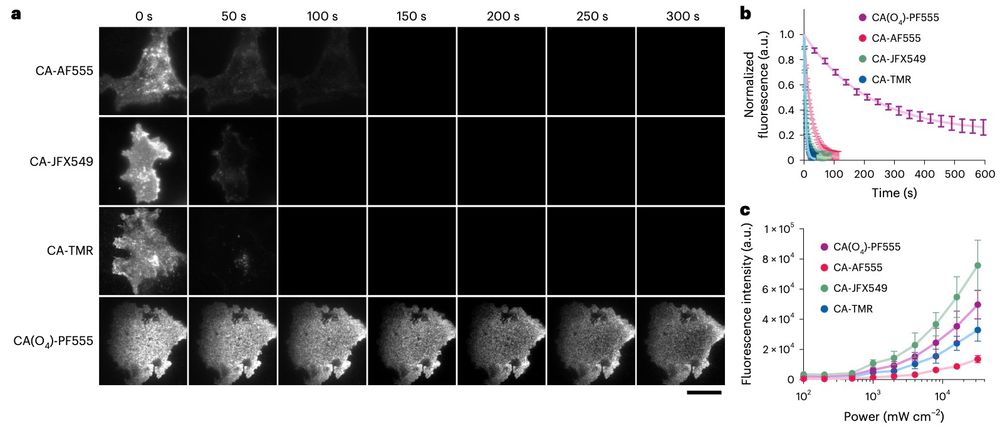
doi.org/10.1038/s415...


