Osterman Ilya
@ostermanilya.bsky.social
Senior Research Associate, microbiologist, biochemist, The Sorek lab, Weizmann Institute of Science
yes, mainly proteobacteria, I think it is correlated with presence of Dam in the genome
November 6, 2025 at 7:19 PM
yes, mainly proteobacteria, I think it is correlated with presence of Dam in the genome
Thank you! It is not very abundant system, in 30k of isolated genomes we have found approximately 300 Metis systems, so 1%. Maybe there are more of them, because I've observed homologs of MisA in other operons with different architecture, maybe they also could be defensive and activated by m6dAmp
November 6, 2025 at 6:44 PM
Thank you! It is not very abundant system, in 30k of isolated genomes we have found approximately 300 Metis systems, so 1%. Maybe there are more of them, because I've observed homologs of MisA in other operons with different architecture, maybe they also could be defensive and activated by m6dAmp
Thank you. I was also very impressed by specificity and also by the fact that there at least two very different binding domains for m6dAMP and maybe there are more....
November 6, 2025 at 10:24 AM
Thank you. I was also very impressed by specificity and also by the fact that there at least two very different binding domains for m6dAMP and maybe there are more....
Thank you, David!
November 6, 2025 at 10:12 AM
Thank you, David!
Thank you Francois:)
November 6, 2025 at 9:43 AM
Thank you Francois:)
Thank you, Aude :) And thank you for the beautiful review — I really liked it! We’ve also found another effector that cleaves NAD⁺ in a different way than the usual NADases and MisA :)
November 6, 2025 at 9:41 AM
Thank you, Aude :) And thank you for the beautiful review — I really liked it! We’ve also found another effector that cleaves NAD⁺ in a different way than the usual NADases and MisA :)
Huge thanks to Rotem Sorek and everyone in the @soreklab.bsky.social — especially @bhurieva.bsky.social and @erezyirmiya.bsky.social. Also to A. Falkovich, M. Itkin, and S. Malitsky.
And special thanks to the very talented Weizmann student Sarit Moses, who started this project during her rotation.
And special thanks to the very talented Weizmann student Sarit Moses, who started this project during her rotation.
November 6, 2025 at 5:00 AM
Huge thanks to Rotem Sorek and everyone in the @soreklab.bsky.social — especially @bhurieva.bsky.social and @erezyirmiya.bsky.social. Also to A. Falkovich, M. Itkin, and S. Malitsky.
And special thanks to the very talented Weizmann student Sarit Moses, who started this project during her rotation.
And special thanks to the very talented Weizmann student Sarit Moses, who started this project during her rotation.
We call this new system Metis, after Greek Titaness of wisdom and deep thought — and fittingly, it’s a wise defender that knows when Methylation becomes a problem
A molecular sensor that detects when the cell’s own DNA has been broken down.
A molecular sensor that detects when the cell’s own DNA has been broken down.
November 6, 2025 at 5:00 AM
We call this new system Metis, after Greek Titaness of wisdom and deep thought — and fittingly, it’s a wise defender that knows when Methylation becomes a problem
A molecular sensor that detects when the cell’s own DNA has been broken down.
A molecular sensor that detects when the cell’s own DNA has been broken down.
Notably, an operon encoding homologs of MisC and MisB was previously shown to defend against phage T7 🔗 linkinghub.elsevier.com/retrieve/pii/S1931-3128(22)00104-4
@francoisrousset.bsky.social @dbikard.bsky.social
@francoisrousset.bsky.social @dbikard.bsky.social
November 6, 2025 at 5:00 AM
Notably, an operon encoding homologs of MisC and MisB was previously shown to defend against phage T7 🔗 linkinghub.elsevier.com/retrieve/pii/S1931-3128(22)00104-4
@francoisrousset.bsky.social @dbikard.bsky.social
@francoisrousset.bsky.social @dbikard.bsky.social
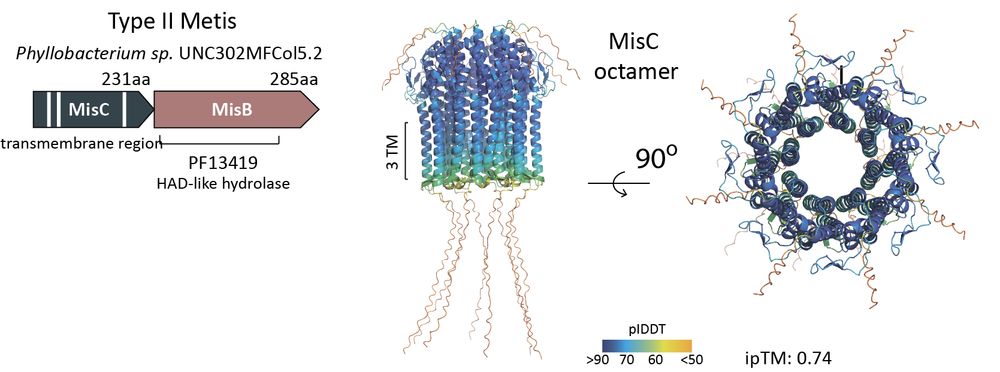
In many bacteria, MisB pairs not with MisA but with MisC, a protein with three transmembrane helices.
Together, they form type II Metis — a defense system also activated by m⁶dAMP.
AF3 predictions suggest that MisC could assemble into an octameric pore across the membrane during activation.
Together, they form type II Metis — a defense system also activated by m⁶dAMP.
AF3 predictions suggest that MisC could assemble into an octameric pore across the membrane during activation.
November 6, 2025 at 5:00 AM
In many bacteria, MisB pairs not with MisA but with MisC, a protein with three transmembrane helices.
Together, they form type II Metis — a defense system also activated by m⁶dAMP.
AF3 predictions suggest that MisC could assemble into an octameric pore across the membrane during activation.
Together, they form type II Metis — a defense system also activated by m⁶dAMP.
AF3 predictions suggest that MisC could assemble into an octameric pore across the membrane during activation.
Metis doesn’t recognize a specific phage protein — it recognizes a consequence of infection: genome degradation.
Phages can escape Metis by avoiding host DNA degradation (for example, through mutation of the DenA endonuclease), but that escape comes at a cost —they lose access to host nucleotides.
Phages can escape Metis by avoiding host DNA degradation (for example, through mutation of the DenA endonuclease), but that escape comes at a cost —they lose access to host nucleotides.

November 6, 2025 at 5:00 AM
Metis doesn’t recognize a specific phage protein — it recognizes a consequence of infection: genome degradation.
Phages can escape Metis by avoiding host DNA degradation (for example, through mutation of the DenA endonuclease), but that escape comes at a cost —they lose access to host nucleotides.
Phages can escape Metis by avoiding host DNA degradation (for example, through mutation of the DenA endonuclease), but that escape comes at a cost —they lose access to host nucleotides.
The second gene, MisB, is an m⁶dAMP phosphatase.
Low levels of m⁶dAMP can appear even without phage infection, during normal DNA repair. MisB “cleans” these traces to prevent premature activation and self-toxicity.
Low levels of m⁶dAMP can appear even without phage infection, during normal DNA repair. MisB “cleans” these traces to prevent premature activation and self-toxicity.

November 6, 2025 at 5:00 AM
The second gene, MisB, is an m⁶dAMP phosphatase.
Low levels of m⁶dAMP can appear even without phage infection, during normal DNA repair. MisB “cleans” these traces to prevent premature activation and self-toxicity.
Low levels of m⁶dAMP can appear even without phage infection, during normal DNA repair. MisB “cleans” these traces to prevent premature activation and self-toxicity.
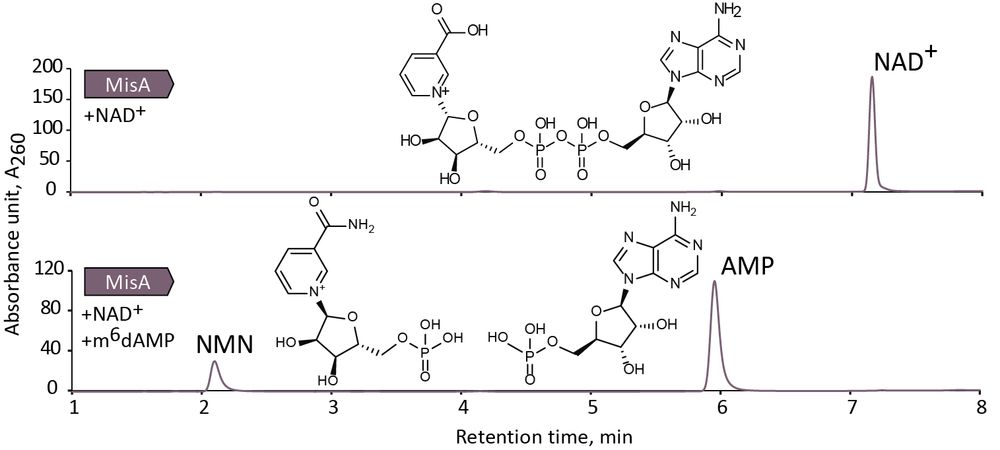
We also found that MisA cuts NAD⁺ in a unique way, producing NMN and AMP — unlike any previously known NAD⁺-depleting defense enzymes.
This might prevent phages from rebuilding NAD⁺ through their usual repair pathways (for example, NARP1 - www.nature.com/articles/s41...).
This might prevent phages from rebuilding NAD⁺ through their usual repair pathways (for example, NARP1 - www.nature.com/articles/s41...).
November 6, 2025 at 5:00 AM
We also found that MisA cuts NAD⁺ in a unique way, producing NMN and AMP — unlike any previously known NAD⁺-depleting defense enzymes.
This might prevent phages from rebuilding NAD⁺ through their usual repair pathways (for example, NARP1 - www.nature.com/articles/s41...).
This might prevent phages from rebuilding NAD⁺ through their usual repair pathways (for example, NARP1 - www.nature.com/articles/s41...).
In this work, we identified a two-gene anti-phage defense system, which we named Metis. Upon accumulation of m⁶dAMP, MisA is activated and rapidly depletes NAD⁺, shutting down the cell and preventing phage replication.

November 6, 2025 at 5:00 AM
In this work, we identified a two-gene anti-phage defense system, which we named Metis. Upon accumulation of m⁶dAMP, MisA is activated and rapidly depletes NAD⁺, shutting down the cell and preventing phage replication.
We found that bacteria can detect the degradation of their own genome — a hallmark of phage infection.
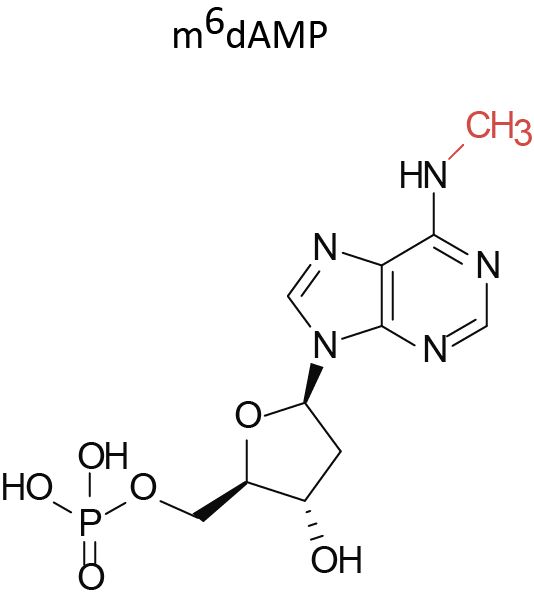
They do it by sensing the accumulation of a modified nucleotide, N⁶-methyl-deoxyadenosine monophosphate (m⁶dAMP) — a signature molecule that appears when bacterial DNA is broken down.
They do it by sensing the accumulation of a modified nucleotide, N⁶-methyl-deoxyadenosine monophosphate (m⁶dAMP) — a signature molecule that appears when bacterial DNA is broken down.
November 6, 2025 at 5:00 AM
We found that bacteria can detect the degradation of their own genome — a hallmark of phage infection.
They do it by sensing the accumulation of a modified nucleotide, N⁶-methyl-deoxyadenosine monophosphate (m⁶dAMP) — a signature molecule that appears when bacterial DNA is broken down.
They do it by sensing the accumulation of a modified nucleotide, N⁶-methyl-deoxyadenosine monophosphate (m⁶dAMP) — a signature molecule that appears when bacterial DNA is broken down.
Phages often degrade host DNA into individual nucleotides to reuse them for building their own genomes.
When this happens, bacterial cells are doomed — but not necessarily defenseless.
When this happens, bacterial cells are doomed — but not necessarily defenseless.
November 6, 2025 at 5:00 AM
Phages often degrade host DNA into individual nucleotides to reuse them for building their own genomes.
When this happens, bacterial cells are doomed — but not necessarily defenseless.
When this happens, bacterial cells are doomed — but not necessarily defenseless.
Reposted by Osterman Ilya
Phage biology: Stuck with dU www.sciencedirect.com/science/arti...
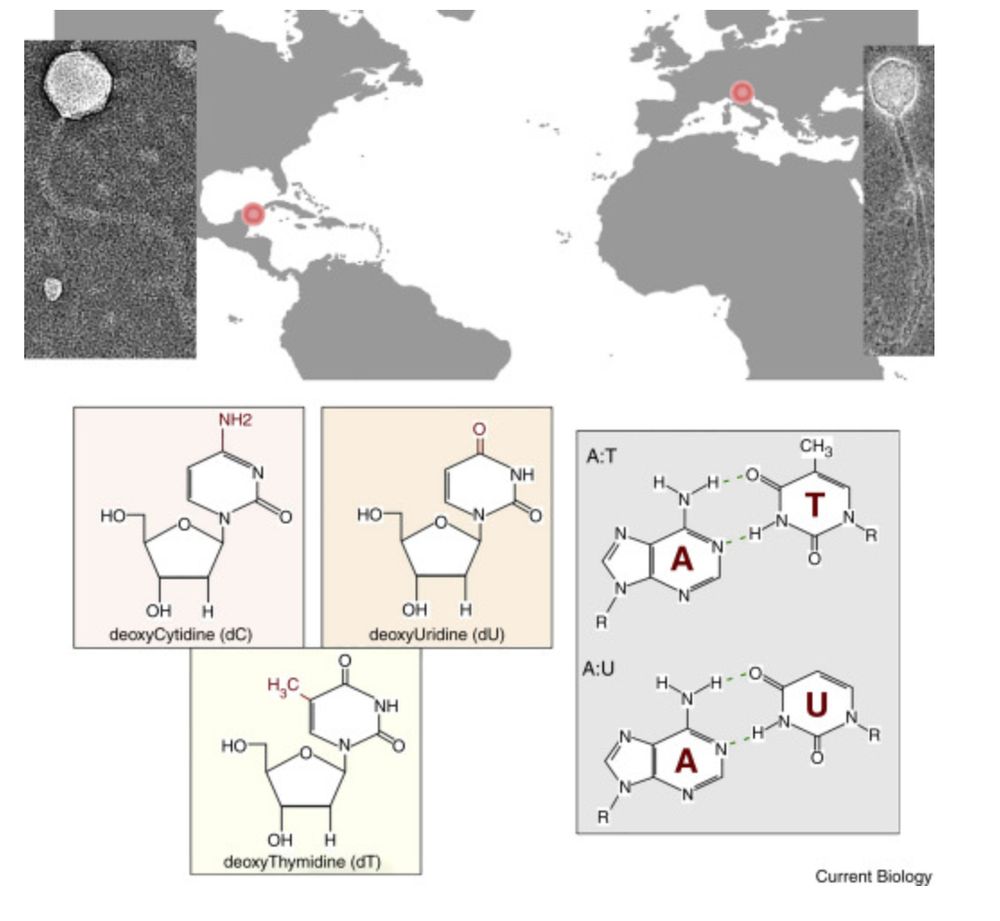
November 4, 2025 at 2:38 PM
Phage biology: Stuck with dU www.sciencedirect.com/science/arti...

