
Lille, France (Bonsai team).
I develop efficient computational methods to analyze massive sequencing data, creating scalable tools for genomics, transcriptomics, and metagenomics.
https://malfoy.github.io/


WELL....

WELL....
WTF is Zoom client doing, mining Bitcoin ?

WTF is Zoom client doing, mining Bitcoin ?


We are able to be order of magnitude more resource-efficient than state of the art and globally scalable!

We are able to be order of magnitude more resource-efficient than state of the art and globally scalable!
movi (move index)
www.biorxiv.org/content/10.1...
SRC (MPHF and fingerprints) www.sciencedirect.com/science/arti...
Fulgor (MPHF and many CDBG tricks)
almob.biomedcentral.com/articles/10....
Themisto (spectral bwt) academic.oup.com/bioinformati...

movi (move index)
www.biorxiv.org/content/10.1...
SRC (MPHF and fingerprints) www.sciencedirect.com/science/arti...
Fulgor (MPHF and many CDBG tricks)
almob.biomedcentral.com/articles/10....
Themisto (spectral bwt) academic.oup.com/bioinformati...
(Be not afraid by the figure)

(Be not afraid by the figure)
2)Successive kmers are very likely to share the exact same origins
3)The amount of distinct "colors" ie read lists grow counterintuitively LINEARLY in an idealized scenario but also quite in practice

2)Successive kmers are very likely to share the exact same origins
3)The amount of distinct "colors" ie read lists grow counterintuitively LINEARLY in an idealized scenario but also quite in practice
To truly optimize compression, we need similar reads on the same strand!
Since this might affect strand-specific analyses, it's an optional feature.

To truly optimize compression, we need similar reads on the same strand!
Since this might affect strand-specific analyses, it's an optional feature.

Here a log scale plot of the compression of a HiFi dataset comparing xz,zstd,bz2,gz with various compression level with and without reordering.
State of the art tools performance are also included for reference.

Here a log scale plot of the compression of a HiFi dataset comparing xz,zstd,bz2,gz with various compression level with and without reordering.
State of the art tools performance are also included for reference.
But the bird-eye view is that we are going to rely on a draft assembly graph to do so.
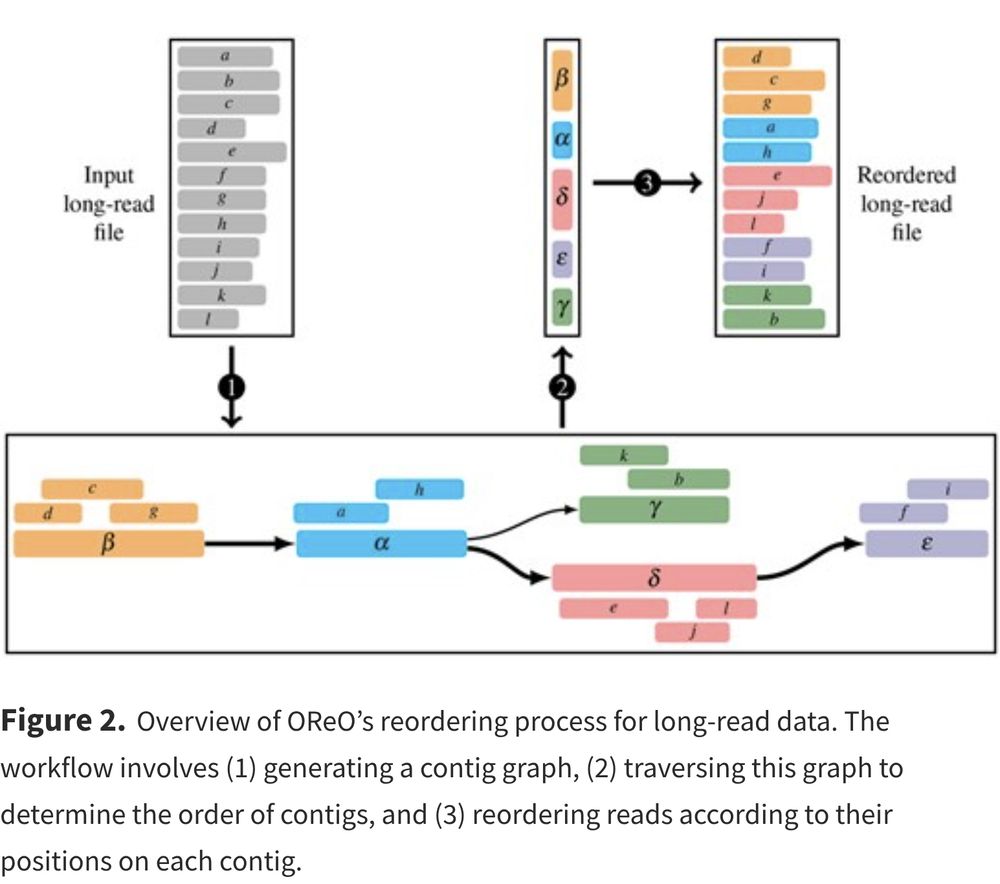
But the bird-eye view is that we are going to rely on a draft assembly graph to do so.



We mean need even faster parser, minimizer iterator or higher density minimizer...

We mean need even faster parser, minimizer iterator or higher density minimizer...
Hence, K2Rmini is basically a accelerated version of back_to_sequence, bringing order of magnitude improvement!

Hence, K2Rmini is basically a accelerated version of back_to_sequence, bringing order of magnitude improvement!
www.ncbi.nlm.nih.gov/sra/docs/sra...

www.ncbi.nlm.nih.gov/sra/docs/sra...
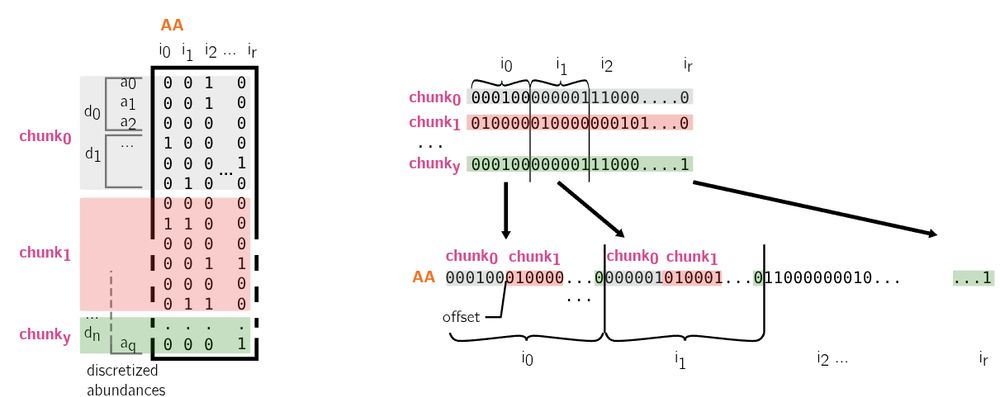

Using sparse structures lets us have many large bloom filters at a very low cost in both time and memory.

Using sparse structures lets us have many large bloom filters at a very low cost in both time and memory.



