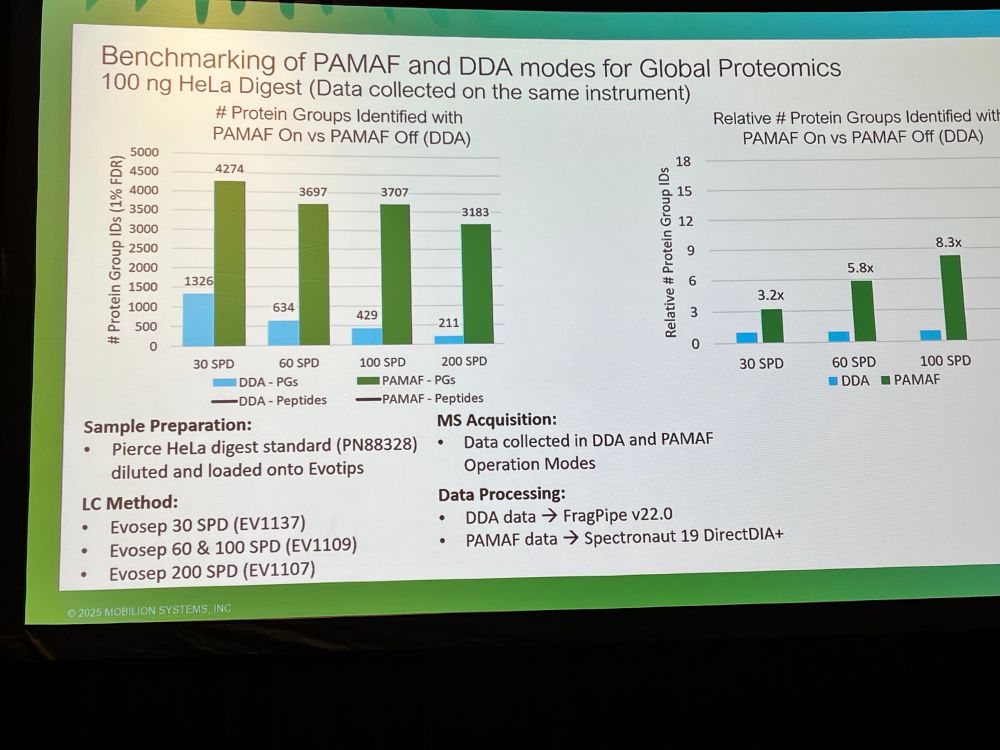

Weather’s going wild, and now your mass spec data’s a mess too?
Coindidence? Nope!
We reveal how weather-driven air pressure fluctuations impact diaPASEF-based high troughput proteomics - and how to fix it!
Check out our new paper! #diaPASEF #Weatheromics
pubs.acs.org/doi/10.1021/...
"trademark DXT for our DIA multiplex tags...advances have been made in DXT multiplexing since ASMS with the number of tags increased from 6 to 11 and with the potential to
increase these to beyond 30"
"trademark DXT for our DIA multiplex tags...advances have been made in DXT multiplexing since ASMS with the number of tags increased from 6 to 11 and with the potential to
increase these to beyond 30"
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Many non-human proteomics studies still search against taxon-filtered FASTAs.
❌ Redundant sequences
❌ Inflated search space
✅ Reference proteomes cut redundancy, improve annotation, and make results comparable.
👉 Time to move beyond taxon filters. #proteomics #massspec #uniprot

Many non-human proteomics studies still search against taxon-filtered FASTAs.
❌ Redundant sequences
❌ Inflated search space
✅ Reference proteomes cut redundancy, improve annotation, and make results comparable.
👉 Time to move beyond taxon filters. #proteomics #massspec #uniprot
This could also work for the Orbitrap Astral.
Bonus: DIAPASEF on Thermo - patentscope.wipo.int/search/en/de...

This could also work for the Orbitrap Astral.
Bonus: DIAPASEF on Thermo - patentscope.wipo.int/search/en/de...

When you run proteomics on non-human species (mouse, rat, macaque, etc.) — which protein FASTA do you prefer?
Taxonomy-filtered UniProt (all entries)
Reference proteome (SwissProt+TrEMBL)
Ensembl/GENCODE
Something else?

When you run proteomics on non-human species (mouse, rat, macaque, etc.) — which protein FASTA do you prefer?
Taxonomy-filtered UniProt (all entries)
Reference proteome (SwissProt+TrEMBL)
Ensembl/GENCODE
Something else?
www.biorxiv.org/content/10.1...
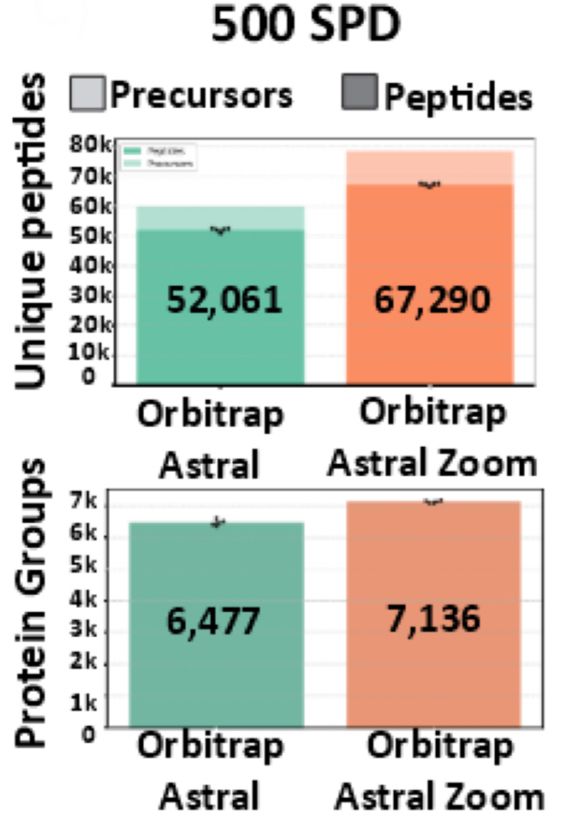
www.biorxiv.org/content/10.1...
github.com/pwilmart/qua...
github.com/pwilmart/qua...
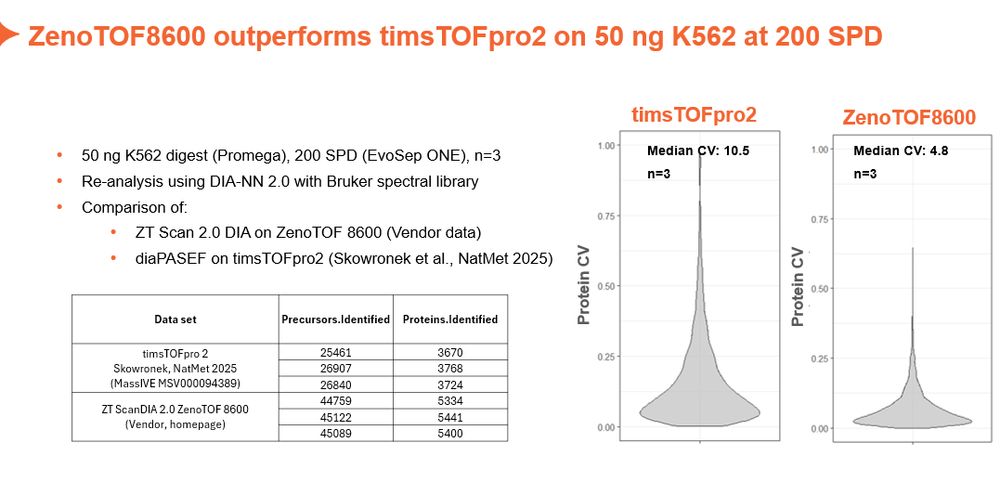
www.nature.com/articles/s41...
www.nature.com/articles/s41...
www.youtube.com/playlist?lis...
www.youtube.com/playlist?lis...
Anyone using WhisperZoom for standard inputs (~500 ng)?
Getting great data on timsTOF Pro Ultra (with ICC2), but repeatedly hit overpressure on Aurora columns - forcing me to discard them.
Anyone else seeing this?
Anyone using WhisperZoom for standard inputs (~500 ng)?
Getting great data on timsTOF Pro Ultra (with ICC2), but repeatedly hit overpressure on Aurora columns - forcing me to discard them.
Anyone else seeing this?
- enhanced ion capacity of the dual-stage TIMS-MX ion funnel
- Athen Ion Processor-equipped timsMetabo, up to 300 Hz PRM
Also QSee software (a nightmare at talks, QC or Qsee?).
As found by Biswapriya Misra
www.bruker.com/en/news-and-...

- enhanced ion capacity of the dual-stage TIMS-MX ion funnel
- Athen Ion Processor-equipped timsMetabo, up to 300 Hz PRM
Also QSee software (a nightmare at talks, QC or Qsee?).
As found by Biswapriya Misra
www.bruker.com/en/news-and-...
timePlex enables time-domain sample multiplexing in LC-MS — boosting proteomics throughput up to 9× with no labels and minimal compromise in quant accuracy.
Combine with plexDIA for 27 samples/run.

timePlex enables time-domain sample multiplexing in LC-MS — boosting proteomics throughput up to 9× with no labels and minimal compromise in quant accuracy.
Combine with plexDIA for 27 samples/run.

www.nature.com/articles/s41...
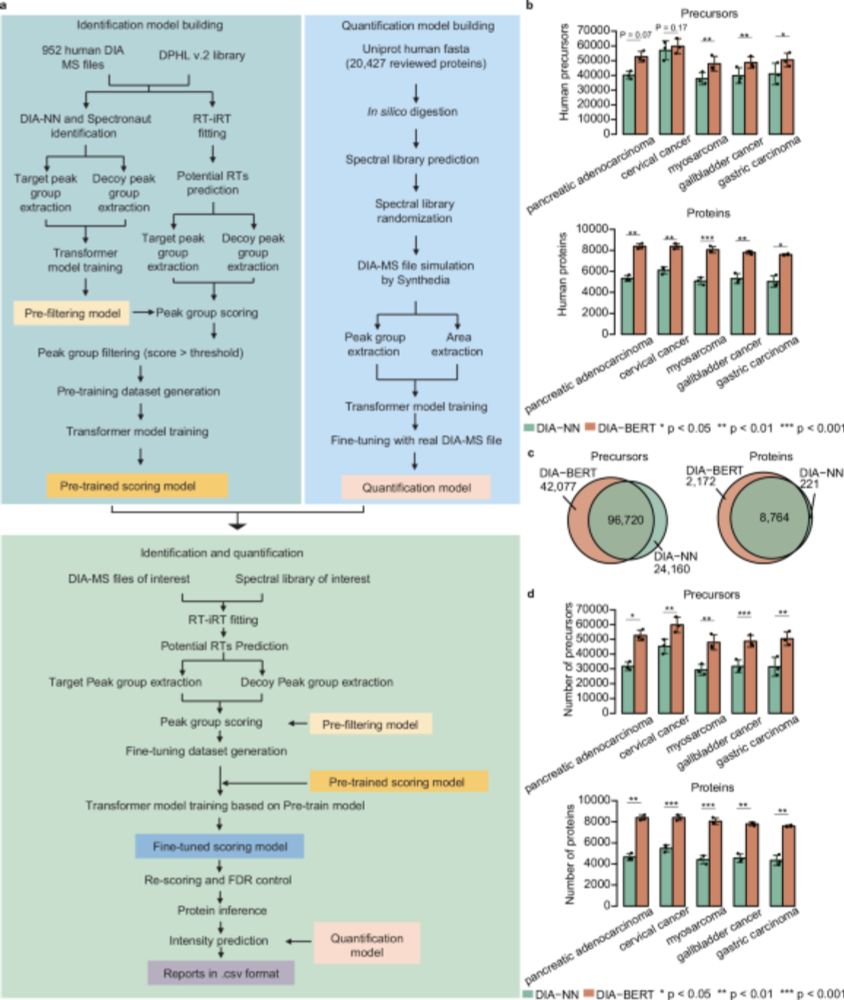
www.nature.com/articles/s41...

---
#proteomics #prot-preprint
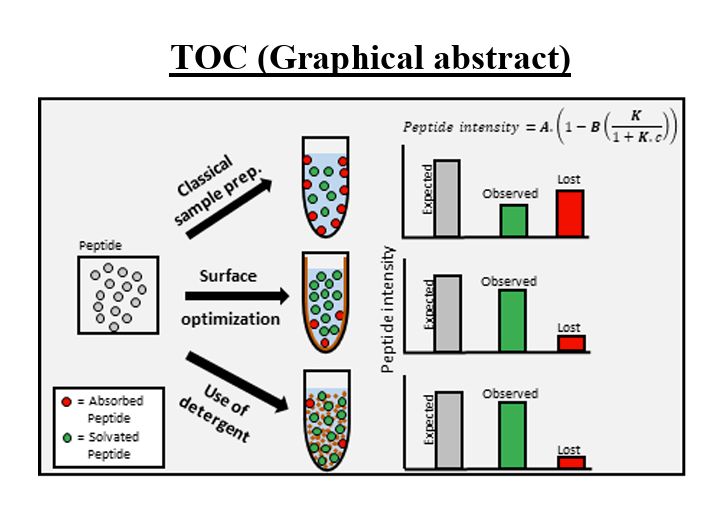
---
#proteomics #prot-preprint
with similar quantitative behavior, providing a new approach to the protein grouping
problem and enabling identification of regulated proteoforms directly from bottom-up data.“
www.biorxiv.org/content/10.1...

with similar quantitative behavior, providing a new approach to the protein grouping
problem and enabling identification of regulated proteoforms directly from bottom-up data.“
www.biorxiv.org/content/10.1...

Does anyone have a clue how to assign multiple MS methods to single run via Bruker HyStar?
pubs.acs.org/doi/10.1021/...

Does anyone have a clue how to assign multiple MS methods to single run via Bruker HyStar?
pubs.acs.org/doi/10.1021/...