@JedNzy. It's about Rubisco and what chaperones are really for. He's on the market, get him while you can. www.biorxiv.org/content/10.1...

@JedNzy. It's about Rubisco and what chaperones are really for. He's on the market, get him while you can. www.biorxiv.org/content/10.1...
Have a look at my recent talk at the @danafarber.bsky.social Targeted Protein Degradation Webinar Series now out on YouTube. Here, I talk about my recent publication (see Blue-torial below)
www.youtube.com/watch?v=Cpg0...
Have a look at my recent talk at the @danafarber.bsky.social Targeted Protein Degradation Webinar Series now out on YouTube. Here, I talk about my recent publication (see Blue-torial below)
www.youtube.com/watch?v=Cpg0...


“𝗨𝗯𝗶𝗥𝗘𝗔𝗗 𝗱𝗲𝗰𝗶𝗽𝗵𝗲𝗿𝘀 𝗽𝗿𝗼𝘁𝗲𝗮𝘀𝗼𝗺𝗮𝗹 𝗱𝗲𝗴𝗿𝗮𝗱𝗮𝘁𝗶𝗼𝗻 𝗰𝗼𝗱𝗲 𝗼𝗳 𝗵𝗼𝗺𝗼𝘁𝘆𝗽𝗶𝗰 𝗮𝗻𝗱 𝗯𝗿𝗮𝗻𝗰𝗵𝗲𝗱 𝗞48 𝗮𝗻𝗱 𝗞63 𝘂𝗯𝗶𝗾𝘂𝗶𝘁𝗶𝗻 𝗰𝗵𝗮𝗶𝗻𝘀”
@cp-molcell.bsky.social
1/8
www.cell.com/molecular-ce...
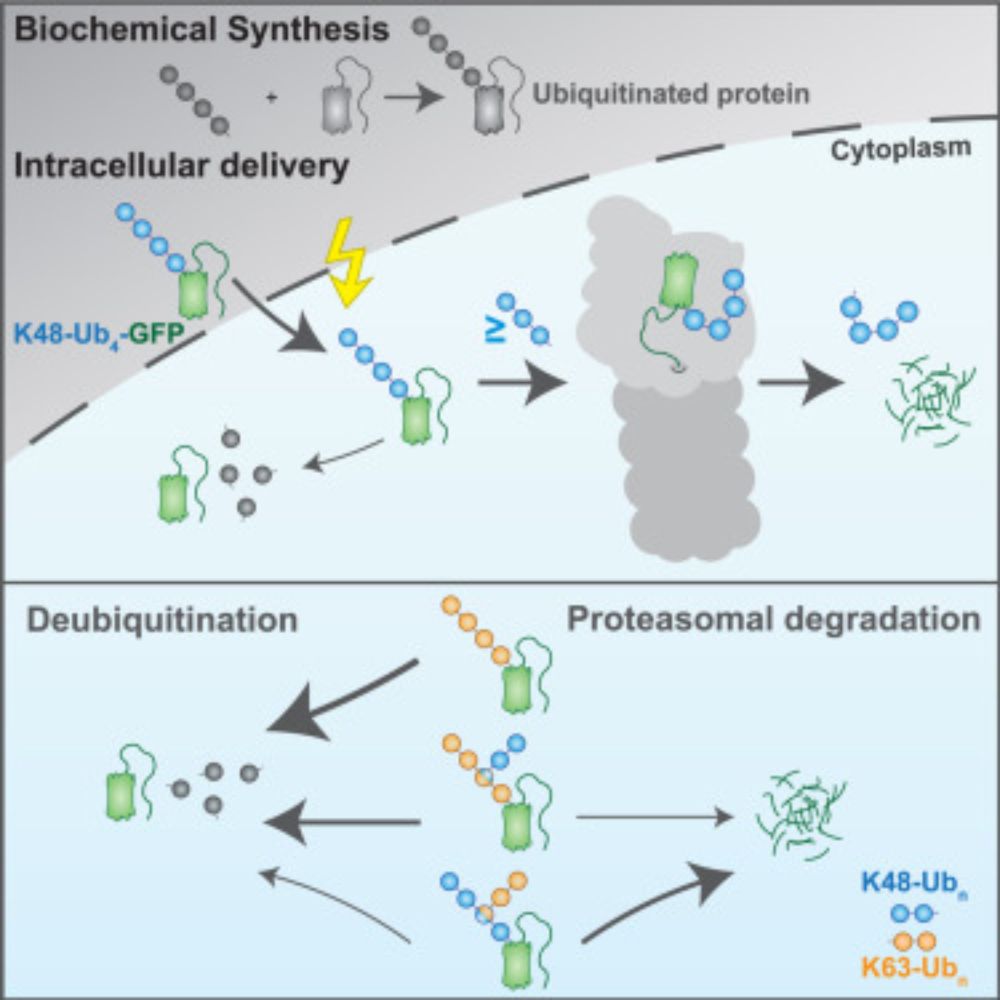
“𝗨𝗯𝗶𝗥𝗘𝗔𝗗 𝗱𝗲𝗰𝗶𝗽𝗵𝗲𝗿𝘀 𝗽𝗿𝗼𝘁𝗲𝗮𝘀𝗼𝗺𝗮𝗹 𝗱𝗲𝗴𝗿𝗮𝗱𝗮𝘁𝗶𝗼𝗻 𝗰𝗼𝗱𝗲 𝗼𝗳 𝗵𝗼𝗺𝗼𝘁𝘆𝗽𝗶𝗰 𝗮𝗻𝗱 𝗯𝗿𝗮𝗻𝗰𝗵𝗲𝗱 𝗞48 𝗮𝗻𝗱 𝗞63 𝘂𝗯𝗶𝗾𝘂𝗶𝘁𝗶𝗻 𝗰𝗵𝗮𝗶𝗻𝘀”
@cp-molcell.bsky.social
1/8
www.cell.com/molecular-ce...
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
🌐 search.foldseek.com
🌐 search.foldseek.com
A lot of insights into RNA Polymerase selectivity at snRNAs promoters !! As these promoters are often used for heterologous expression of RNAs (ie guide RNAs), you better know how they work
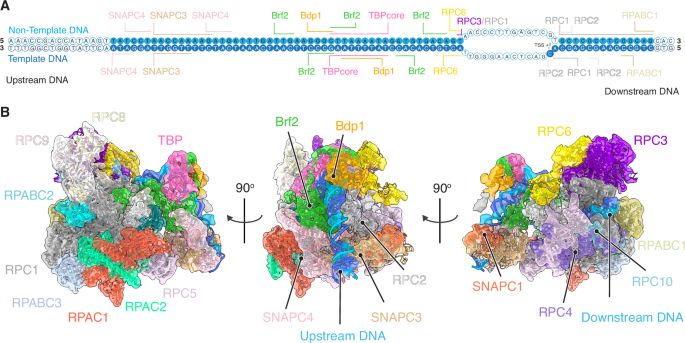
A lot of insights into RNA Polymerase selectivity at snRNAs promoters !! As these promoters are often used for heterologous expression of RNAs (ie guide RNAs), you better know how they work
Read more about the role here:
www.nature.com/naturecareer...
Apply by 30 DEC
#ScienceJobs #CambridgeJobs #PostdocJobs #GenomeEngineering

Read more about the role here:
www.nature.com/naturecareer...
Apply by 30 DEC
#ScienceJobs #CambridgeJobs #PostdocJobs #GenomeEngineering
Vivian Wu, Svetlana Shabalina, Hyunbin Lee, Grace Nye, Eugene Koonin, Alex Gao, Rhiju Das, Wah Chiu

Vivian Wu, Svetlana Shabalina, Hyunbin Lee, Grace Nye, Eugene Koonin, Alex Gao, Rhiju Das, Wah Chiu
Cool collaboration with the Müller and Eustermann Labs @embl.org !
academic.oup.com/nar/advance-...
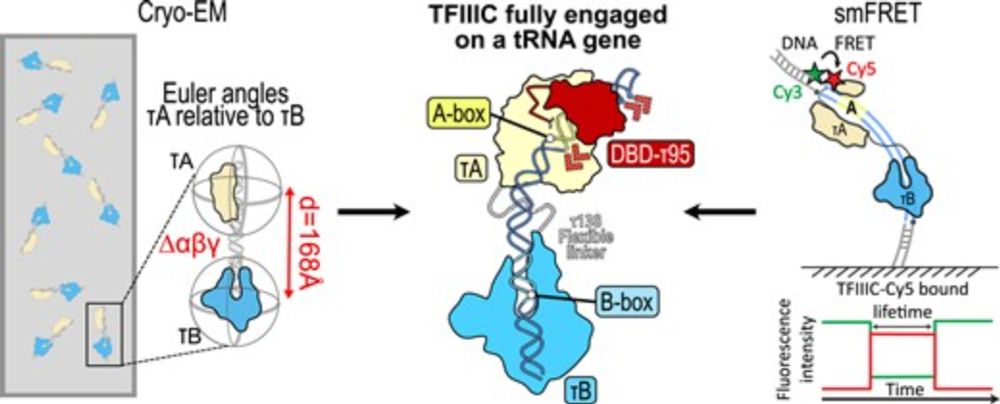
Cool collaboration with the Müller and Eustermann Labs @embl.org !
academic.oup.com/nar/advance-...

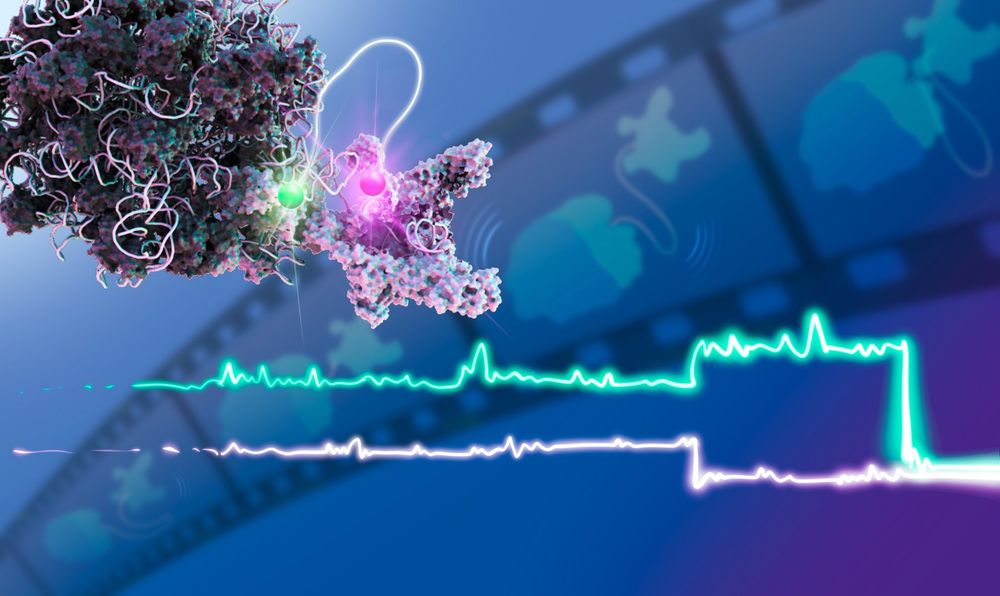
We provide a real-time movie on how #transcription and #translation cooperate using #single-molecule FM: we find long-range #ribosome/ RNAP communication mediated by #RNA looping!
@embl.org #RNAbiology #RNASky
t.co/a24Xxcxdo0
We provide a real-time movie on how #transcription and #translation cooperate using #single-molecule FM: we find long-range #ribosome/ RNAP communication mediated by #RNA looping!
@embl.org #RNAbiology #RNASky
t.co/a24Xxcxdo0
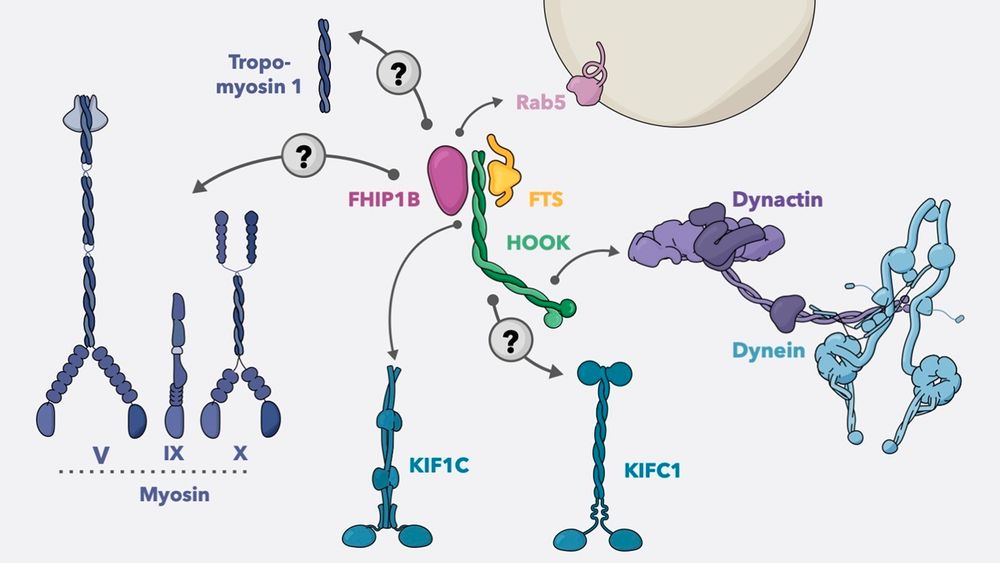
Link: www.nature.com/articles/s41... (1/n)

Link: www.nature.com/articles/s41... (1/n)

