
lichen symbiosis | metagenomics | evolution |
Previously at @thesainsburylab.bsky.social. PhD from @ualberta.bsky.social
Personal website: metalichen.github.io
#StandWithUkraine
she/her




sagdb.uni-goettingen.de/detailedList...

sagdb.uni-goettingen.de/detailedList...
doi.org/10.1111/nph.70728

doi.org/10.1111/nph.70728






tinyurl.com/2w7wwubm

tinyurl.com/2w7wwubm




www.biorxiv.org/content/10.1...
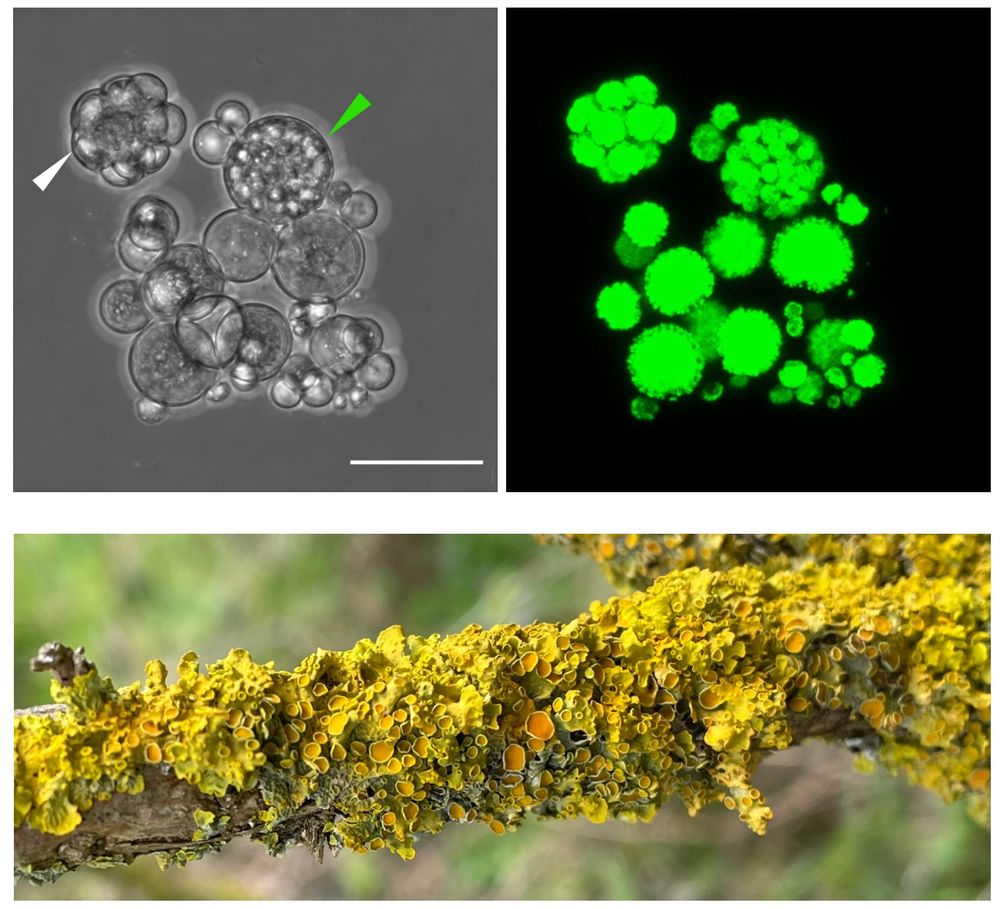
www.biorxiv.org/content/10.1...
#ecfg17

#ecfg17
#ecfg17

#ecfg17
#ecfg17

#ecfg17
#ecfg17

#ecfg17
#ecfg17

#ecfg17
#ecfg17

#ecfg17


