Shuming Liu (MIT), explores the use of IGME non-Markovian model to study chromatin folding in collaboration with Bin Zhang’s group at MIT. One interesting finding: memory kernels decay more slowly in nucleosome
condensates than in free solution.

Shuming Liu (MIT), explores the use of IGME non-Markovian model to study chromatin folding in collaboration with Bin Zhang’s group at MIT. One interesting finding: memory kernels decay more slowly in nucleosome
condensates than in free solution.
▶️ mapped connectivity of nucleosomes in S. cerevisiae
▶️ chromatin is predominantly organized into regularly spaced nucleosome arrays
▶️ metaphase chromosomes packed by arrayed cohesin hubs
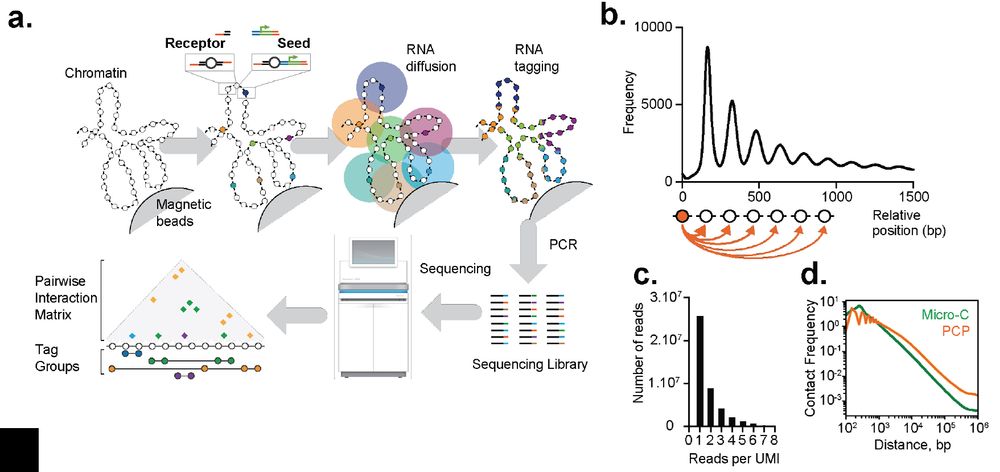
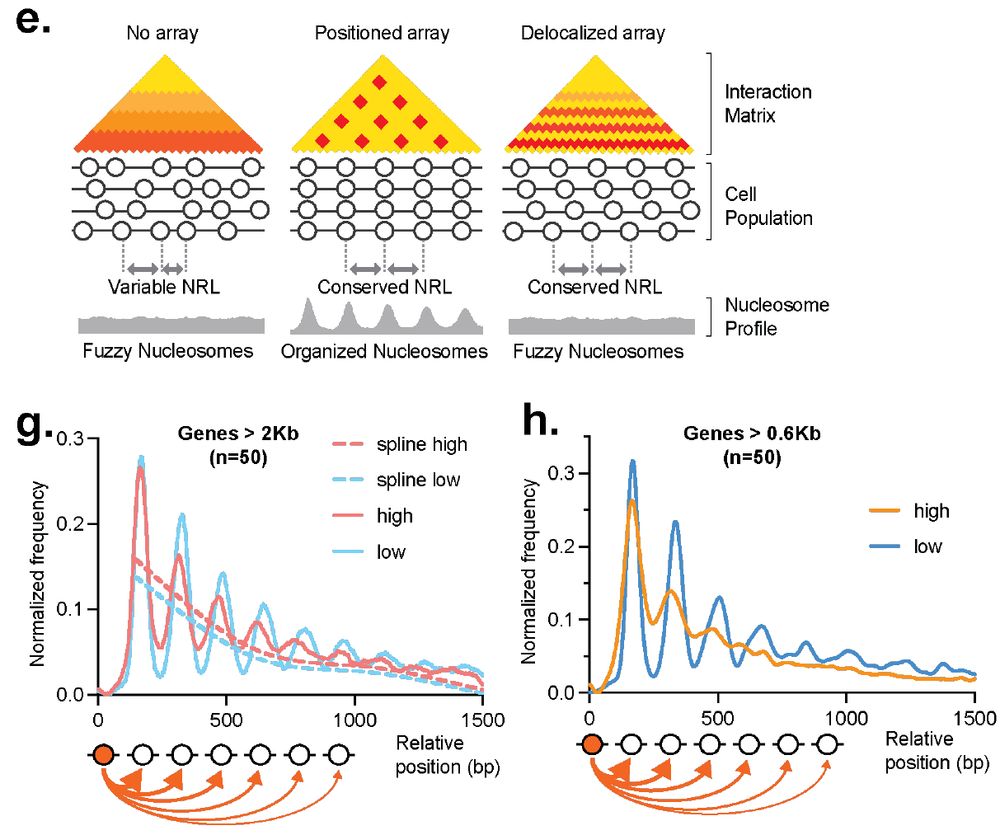
▶️ mapped connectivity of nucleosomes in S. cerevisiae
▶️ chromatin is predominantly organized into regularly spaced nucleosome arrays
▶️ metaphase chromosomes packed by arrayed cohesin hubs
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...

