
https://mdpoleto.github.io/
Image was powered by the one and only MolecularNodes (thx @bradyajohnston.bsky.social) while I was playing around with surface transparency 😁
#chemsky #compchem

Image was powered by the one and only MolecularNodes (thx @bradyajohnston.bsky.social) while I was playing around with surface transparency 😁
#chemsky #compchem


www.epe.gov.br/pt/publicaco...

www.epe.gov.br/pt/publicaco...
We extended the Drude polarizable force field to model PET polymer and demonstrated interesting applications in modeling protein-PET interactions.
Feel free to check it out! DMs are open for feedback 😁

We extended the Drude polarizable force field to model PET polymer and demonstrated interesting applications in modeling protein-PET interactions.
Feel free to check it out! DMs are open for feedback 😁
The past 4 years at Virginia Tech in @justinlemkulvt.bsky.social's lab have been an incredible joyride. I've had the privilege of working with an amazing mentor and friends who have truly shaped this incredible experience♥️
Exciting things to come!😁

The past 4 years at Virginia Tech in @justinlemkulvt.bsky.social's lab have been an incredible joyride. I've had the privilege of working with an amazing mentor and friends who have truly shaped this incredible experience♥️
Exciting things to come!😁


Upon K+ binding at S3, the electric field at the site increases, somewhat trapping the bound ion there.

Upon K+ binding at S3, the electric field at the site increases, somewhat trapping the bound ion there.

(Deoxy)ribose puckering analyses show distinct patterns as well: mostly C2' for telomeric GQ and mixed C2'/C3' conformations for TERRA GQ.

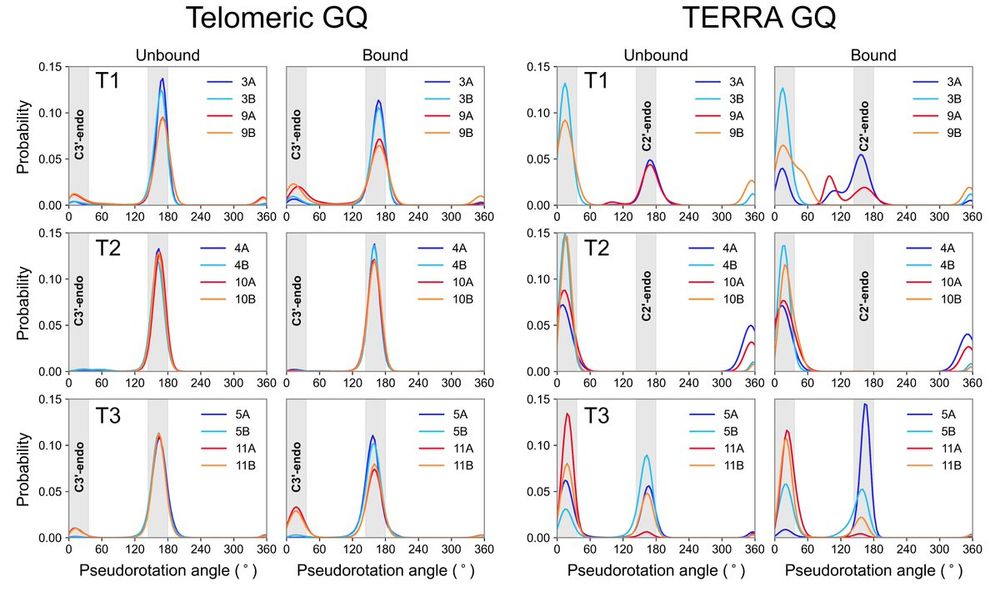
(Deoxy)ribose puckering analyses show distinct patterns as well: mostly C2' for telomeric GQ and mixed C2'/C3' conformations for TERRA GQ.

It has been a 2-year long journey that I am happy to share with everyone. See you all! 😉

It has been a 2-year long journey that I am happy to share with everyone. See you all! 😉


In ATOM MODE, the position of an atom (or COG of multiple atoms) is tracked throughout a trajectory; In our paper, we describe the electric field sensed by a benzene ligand exerted by its receptor (lysozyme) using the Drude polarizable force field.

In ATOM MODE, the position of an atom (or COG of multiple atoms) is tracked throughout a trajectory; In our paper, we describe the electric field sensed by a benzene ligand exerted by its receptor (lysozyme) using the Drude polarizable force field.

Me: "I tried to refine some parameters..."
😮💨
Me: "I tried to refine some parameters..."
😮💨

et. al for their amazing ColabFold!)

et. al for their amazing ColabFold!)
Time for a thread! 🧵👇
#LatinXChem #LXChemComp #Comp166

Time for a thread! 🧵👇
#LatinXChem #LXChemComp #Comp166
I've always been a fan of innovative peer-reviewing strategies and I am happy to be part of a team that is pursuing that.
Looking forward for great science and tons of learning!

I've always been a fan of innovative peer-reviewing strategies and I am happy to be part of a team that is pursuing that.
Looking forward for great science and tons of learning!


