together with Sahar Hani (supported by Arianna Ravera, UNIL’s image analysis expert!), who uncovered hierarchical meta-loop formation. 👏
together with Sahar Hani (supported by Arianna Ravera, UNIL’s image analysis expert!), who uncovered hierarchical meta-loop formation. 👏


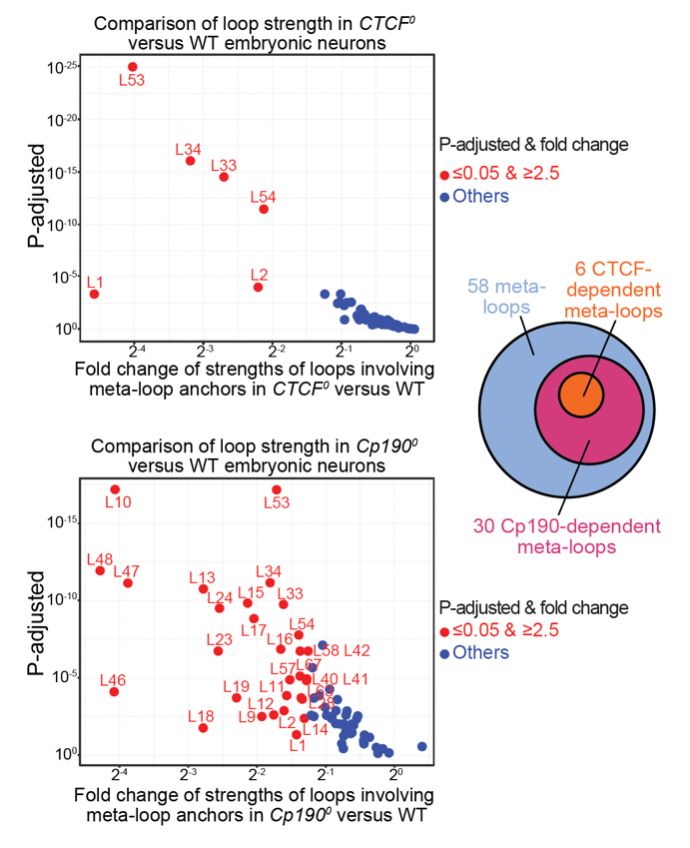
Most meta-domains contain 2 or more meta-loops, with some of these forming earlier during neurogenesis than the other(s) in the same meta-domain.
We found that in many meta-domains, specific meta-loops (often the earlier forming ones) nucleate others.

Most meta-domains contain 2 or more meta-loops, with some of these forming earlier during neurogenesis than the other(s) in the same meta-domain.
We found that in many meta-domains, specific meta-loops (often the earlier forming ones) nucleate others.

2 meta-domains we studied contain a meta-loop connecting a neuronal gene promoter to a distant intergenic element. Disrupting the meta-domain decreased neuronal gene transcription by 2-to-3-fold.
2 meta-domains we studied contain a meta-loop connecting a neuronal gene promoter to a distant intergenic element. Disrupting the meta-domain decreased neuronal gene transcription by 2-to-3-fold.
Within meta-domains, meta-loops connect accessible chromatin peaks present at promoters of specific neuronal genes or in non-coding intergenic DNA.

Within meta-domains, meta-loops connect accessible chromatin peaks present at promoters of specific neuronal genes or in non-coding intergenic DNA.
Thank you @andersshansen.bsky.social and Marcelo Nollmann for including our review in the 2025 Genome Architecture & Expression edition of COGEDE 😀
Thank you @andersshansen.bsky.social and Marcelo Nollmann for including our review in the 2025 Genome Architecture & Expression edition of COGEDE 😀
Open access paper: www.sciencedirect.com/science/arti... Step-by-step protocol: www.sciencedirect.com/science/arti...

Open access paper: www.sciencedirect.com/science/arti... Step-by-step protocol: www.sciencedirect.com/science/arti...
Not all insulator-binding protein sites act as enhancer-blockers.
Not all TAD boundaries act as enhancer-blockers.
Both a CTCF motif and its surrounding sequence context are essential for enhancer-blocking.

Not all insulator-binding protein sites act as enhancer-blockers.
Not all TAD boundaries act as enhancer-blockers.
Both a CTCF motif and its surrounding sequence context are essential for enhancer-blocking.

(1) It identifies biologically meaningful insulators in genomic DNA (both known and novel).

(1) It identifies biologically meaningful insulators in genomic DNA (both known and novel).


