
inspirehep.net/literature?q...
inspirehep.net/literature/1...
Nice little milestone for us all to celebrate, @lukasheinrich.com, @giordonstark.com, @kylecranmer.bsky.social. Cheers!

inspirehep.net/literature?q...
inspirehep.net/literature/1...
Nice little milestone for us all to celebrate, @lukasheinrich.com, @giordonstark.com, @kylecranmer.bsky.social. Cheers!

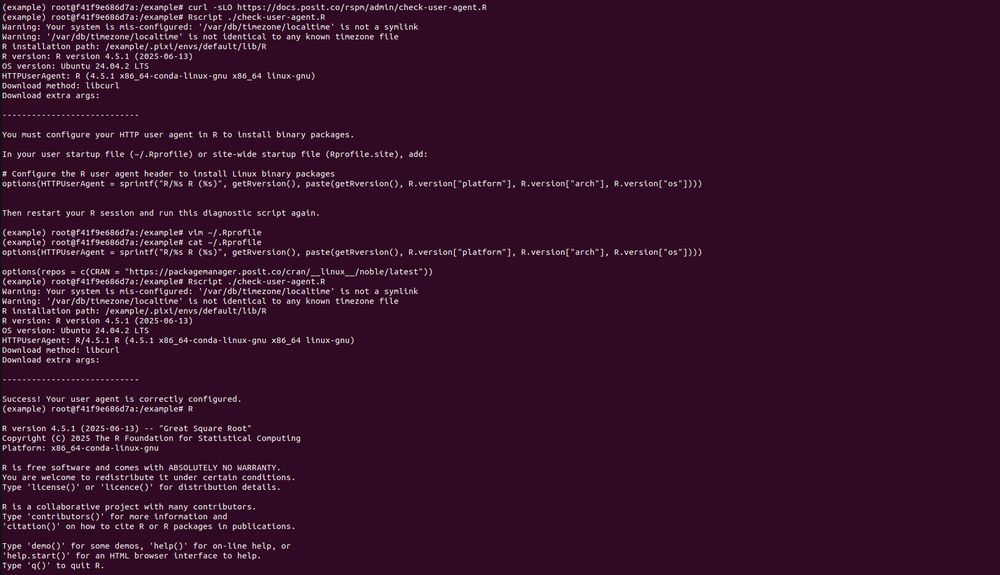
```
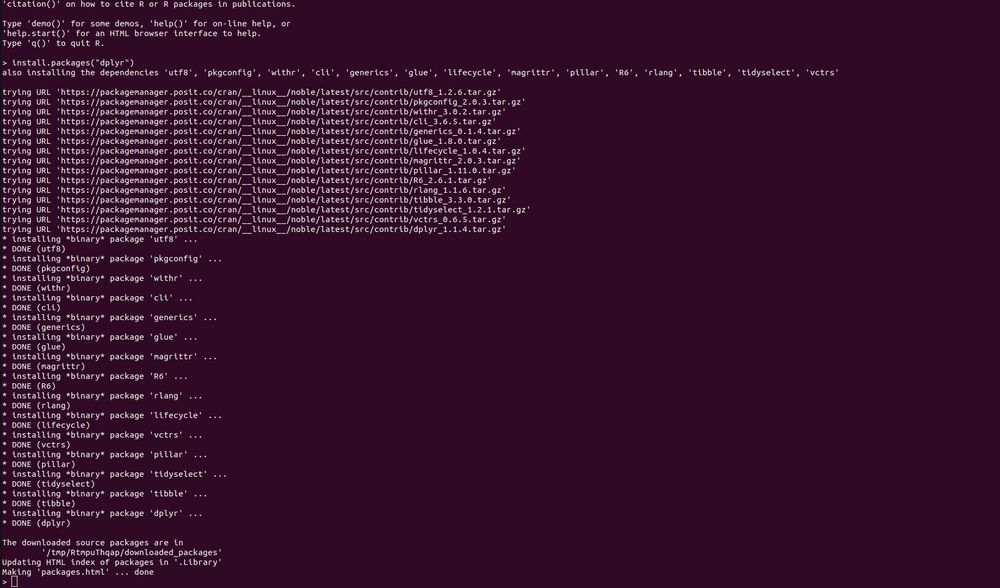
pak::pkg_install("dplyr")
```
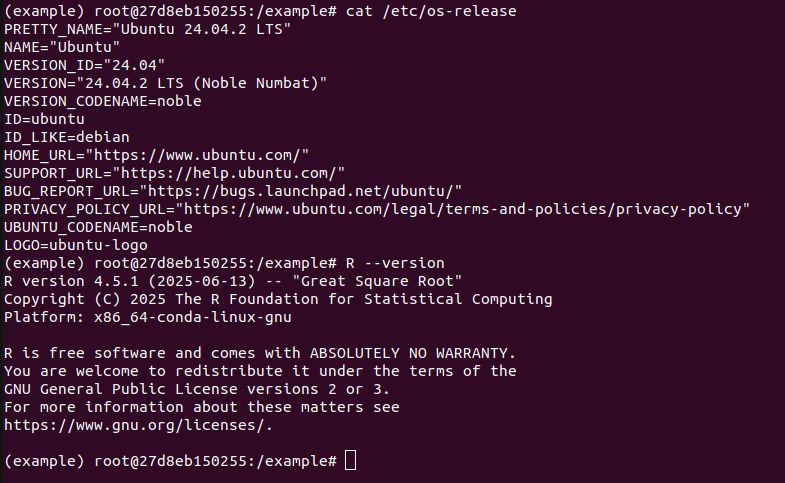
do? It builds 15 packages from source. This is for a Ubuntu 22.04 x86 Linux container with R v4.5.x. So pak is not getting built distributions, because they don't exist. This is not a solution.
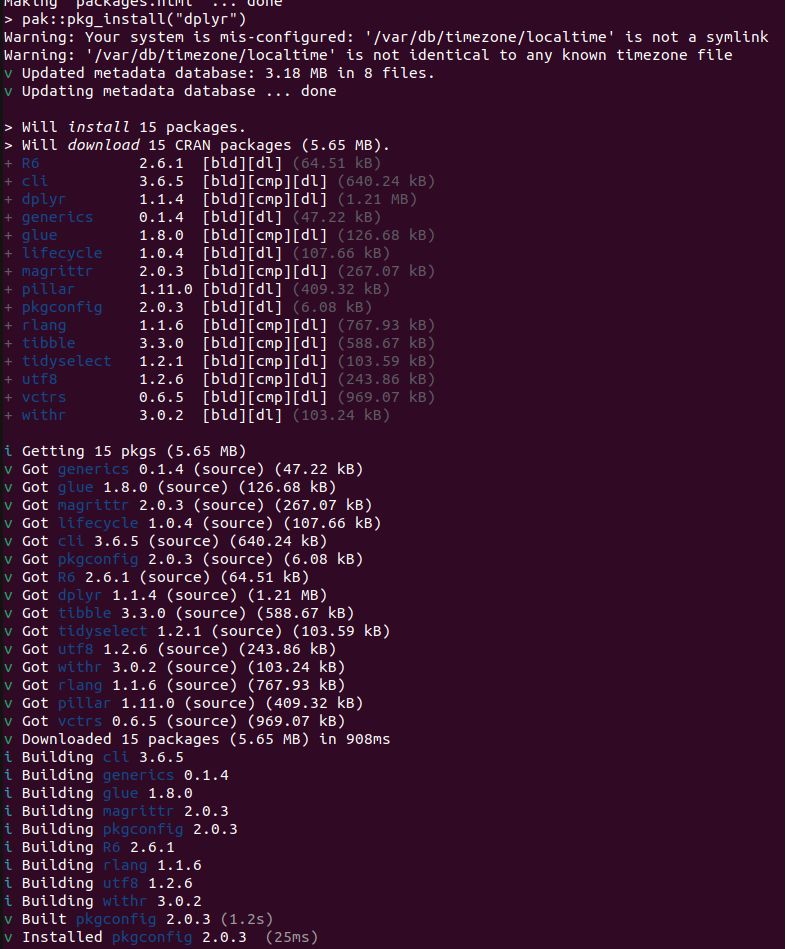
```
pak::pkg_install("dplyr")
```
do? It builds 15 packages from source. This is for a Ubuntu 22.04 x86 Linux container with R v4.5.x. So pak is not getting built distributions, because they don't exist. This is not a solution.
```
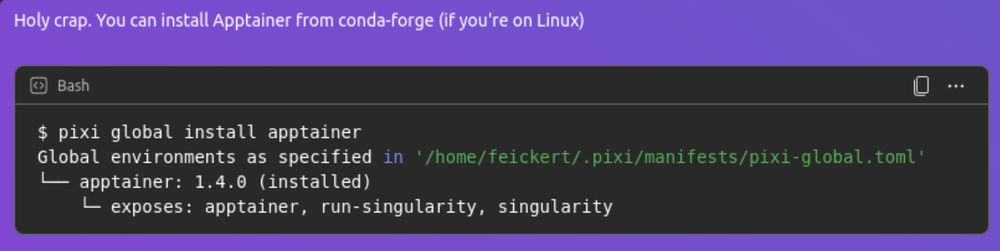
pixi workspace channel add bioconda
```
now means that your Pixi manifest "knows" about every package on Bioconda and can solve against it _and_ conda-forge.

```
pixi workspace channel add bioconda
```
now means that your Pixi manifest "knows" about every package on Bioconda and can solve against it _and_ conda-forge.
Short answer: Run `pixi list`
This will show multiple columns of information including "source", which shows things are coming from conda-forge. conda-forge.org/packages/

Short answer: Run `pixi list`
This will show multiple columns of information including "source", which shows things are coming from conda-forge. conda-forge.org/packages/
* packagemanager.posit.co/client/#/rep...
* packagemanager.posit.co/client/#/rep...
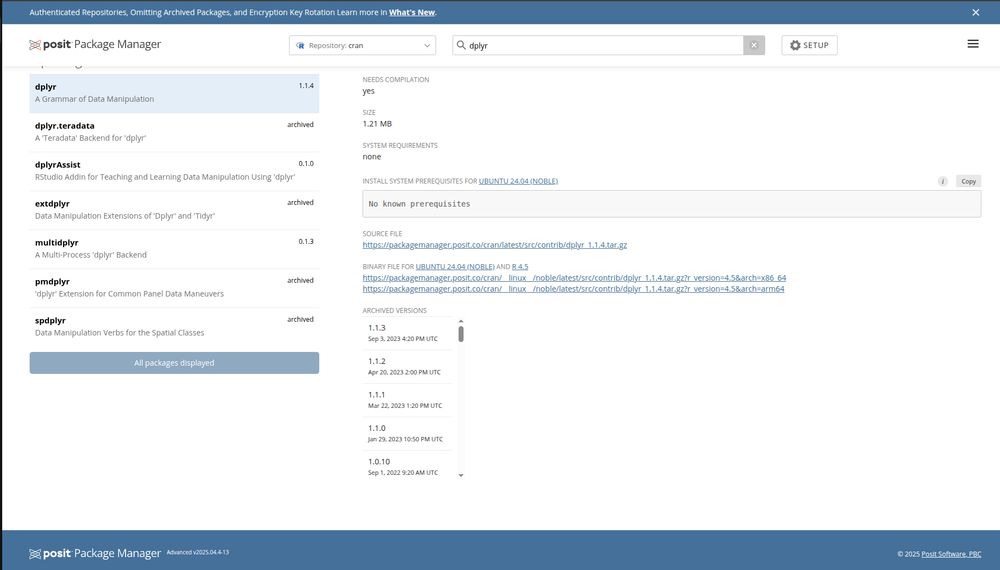


* packagemanager.posit.co/client/#/rep...
* packagemanager.posit.co/client/#/rep...








(Credit to my colleague Jim Pivarski whose talk I took the image from: indico.jlab.org/event/505/co...)
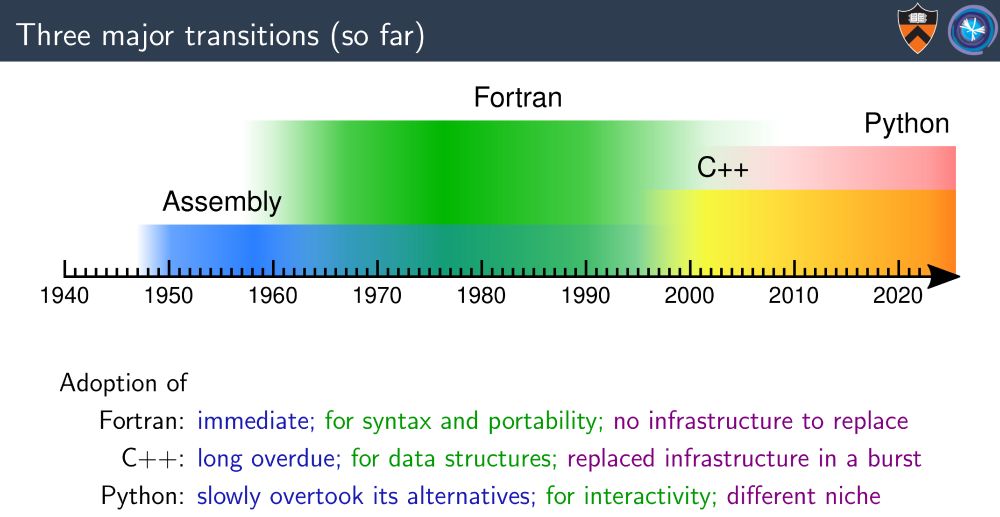
(Credit to my colleague Jim Pivarski whose talk I took the image from: indico.jlab.org/event/505/co...)
github.com/mwaskom/seab...

github.com/mwaskom/seab...













