📄 Check it out here: doi.org/10.1101/2025...
👇 Highlights below

📄 Check it out here: doi.org/10.1101/2025...
👇 Highlights below
simplystatistics.org/posts/2024-1...

simplystatistics.org/posts/2024-1...
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
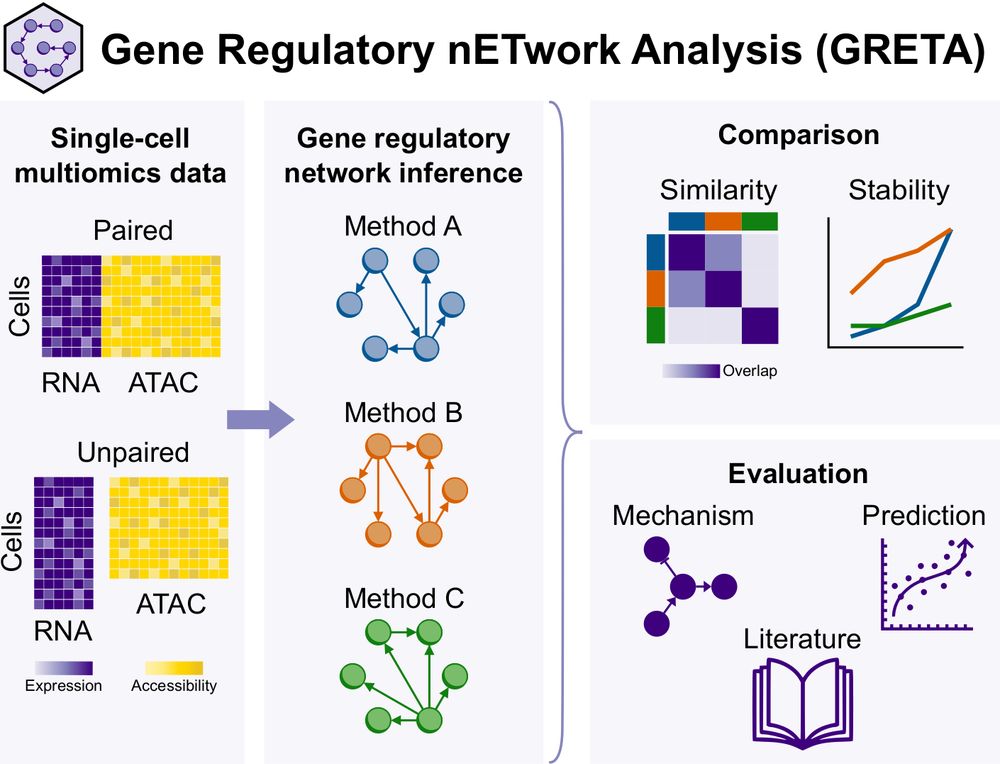
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
logconference.org what a great resource!
logconference.org what a great resource!
www.nature.com/articles/s41...
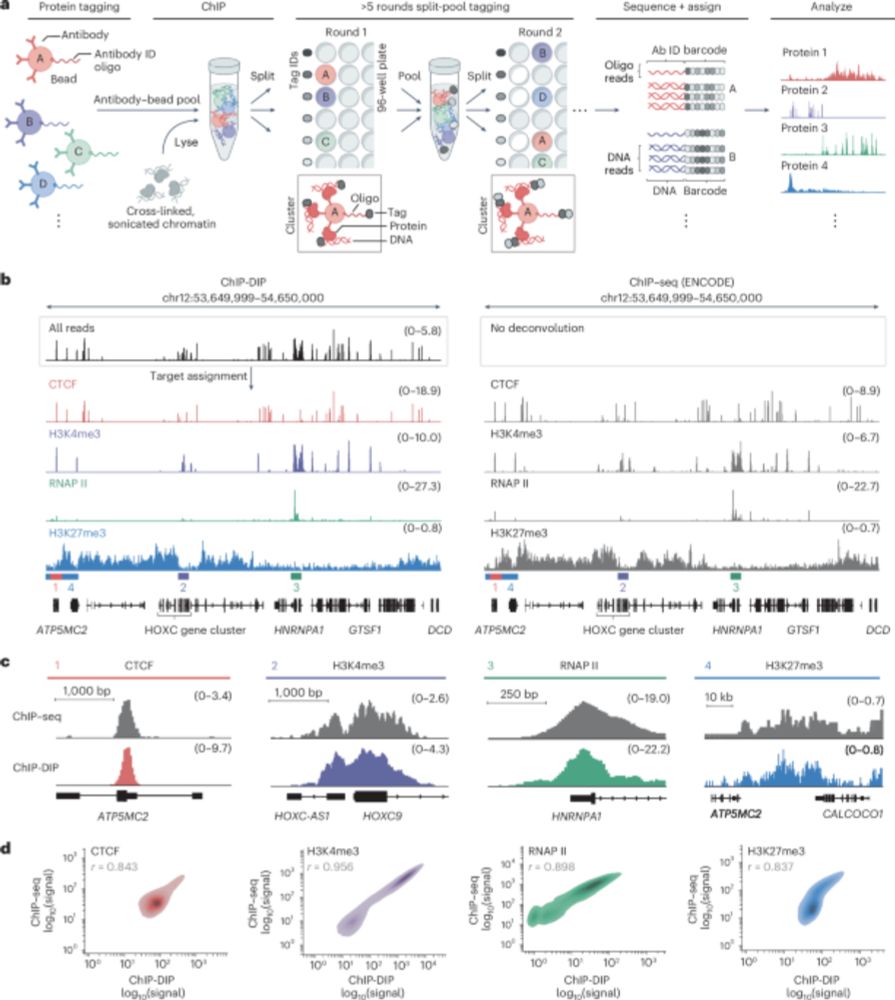
www.nature.com/articles/s41...

