
Microbiome researcher
Based in Brisbane
The biggest change is internal, using a much better approach to saving and loading models (thus removing the dependency on particular versions of scikit-learn)
A few other bugfixes were included
github.com/BigDataBiolo...

The biggest change is internal, using a much better approach to saving and loading models (thus removing the dependency on particular versions of scikit-learn)
A few other bugfixes were included
github.com/BigDataBiolo...
We’re thrilled to share that Gene Tyson, @benjwoodcroft.bsky.social and @luispedrocoelho.bsky.social have once again been named Highly Cited Researchers for 2025 by Clarivate!
#HighlyCited2025
We’re thrilled to share that Gene Tyson, @benjwoodcroft.bsky.social and @luispedrocoelho.bsky.social have once again been named Highly Cited Researchers for 2025 by Clarivate!
#HighlyCited2025
proGenomes4: providing 2 million accurately and consistently annotated high-quality prokaryotic genomes: academic.oup.com/nar/article/...

proGenomes4: providing 2 million accurately and consistently annotated high-quality prokaryotic genomes: academic.oup.com/nar/article/...

Credit to my team and collaborators who have made all of this possible and happy to also see that the Centre for Microbiome Research has 3 highly cited researchers (myself, Ben Woodcroft and Gene Tyson)!
#HighlyCited2025

Credit to my team and collaborators who have made all of this possible and happy to also see that the Centre for Microbiome Research has 3 highly cited researchers (myself, Ben Woodcroft and Gene Tyson)!
#HighlyCited2025
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Huge thanks to @oskolkov.bsky.social , @aroneys.bsky.social and all the amazing participants who joined us this week. 🙌
We wish you all the best with your research projects!



Huge thanks to @oskolkov.bsky.social , @aroneys.bsky.social and all the amazing participants who joined us this week. 🙌
We wish you all the best with your research projects!
@bigdatabiology.bsky.social lab in Shanghai see the light! The work includes my three favorite things research-wise: 🦠 microbiome, 🧬 long-reads, and 🐶 dogs.
See our new preprint:
www.biorxiv.org/content/10.1...

@bigdatabiology.bsky.social lab in Shanghai see the light! The work includes my three favorite things research-wise: 🦠 microbiome, 🧬 long-reads, and 🐶 dogs.
See our new preprint:
www.biorxiv.org/content/10.1...
Using ONT+Illumina, we get better MAGs than to corresponding species representative in public databases
doi.org/10.1101/2025...

Using ONT+Illumina, we get better MAGs than to corresponding species representative in public databases
doi.org/10.1101/2025...
We’re kicking things off with Session 2: Functional insights into microbial communities 🧬
🔹 First talk:
“Small proteins of the human microbiome and beyond”
by Luis Pedro Coelho – Queensland University of Technology
Stay tuned for more exciting science!




We’re kicking things off with Session 2: Functional insights into microbial communities 🧬
🔹 First talk:
“Small proteins of the human microbiome and beyond”
by Luis Pedro Coelho – Queensland University of Technology
Stay tuned for more exciting science!
#GloMiNe2025 Atlantic event closing talks:
Viral diversity in the human microbiome
by Curtis Huttenhower (@hutlab.bsky.social)
and
Cataloging the global microbiome. Very big data for very small things
by @luispedrocoelho.bsky.social (@bigdatabiology.bsky.social)
9/n

#GloMiNe2025 Atlantic event closing talks:
Viral diversity in the human microbiome
by Curtis Huttenhower (@hutlab.bsky.social)
and
Cataloging the global microbiome. Very big data for very small things
by @luispedrocoelho.bsky.social (@bigdatabiology.bsky.social)
9/n
If you need to survive on having many citations to your tools, it is better if your tool gives users lots of results so that they cite it!
open.substack.com/pub/luispedr...
If you need to survive on having many citations to your tools, it is better if your tool gives users lots of results so that they cite it!
open.substack.com/pub/luispedr...
Europe doubles down on research competitiveness with a major boost to #HorizonEurope:
€95.5 billion foreseen for 2021-2027
💰💰💰💰💰💰💰💰💰
€175 billion proposed for 2028-2034
💰💰💰💰💰💰💰💰💰💰💰💰💰💰💰💰💰💰
Europe doubles down on research competitiveness with a major boost to #HorizonEurope:
€95.5 billion foreseen for 2021-2027
💰💰💰💰💰💰💰💰💰
€175 billion proposed for 2028-2034
💰💰💰💰💰💰💰💰💰💰💰💰💰💰💰💰💰💰
- predict protein-protein interactions, operon structure, and protein function
- infer phenotypic traits
- design synthetic genomes with desired properties
@macwiatrak.bsky.social @mariabrbic.bsky.social
@andresfloto.bsky.social
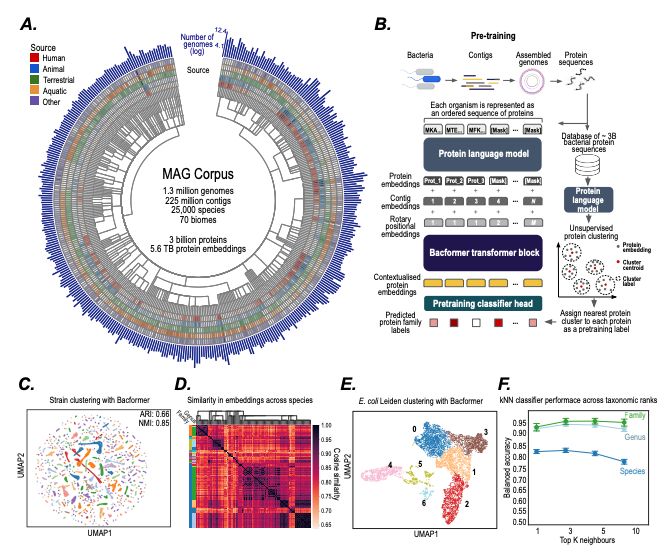
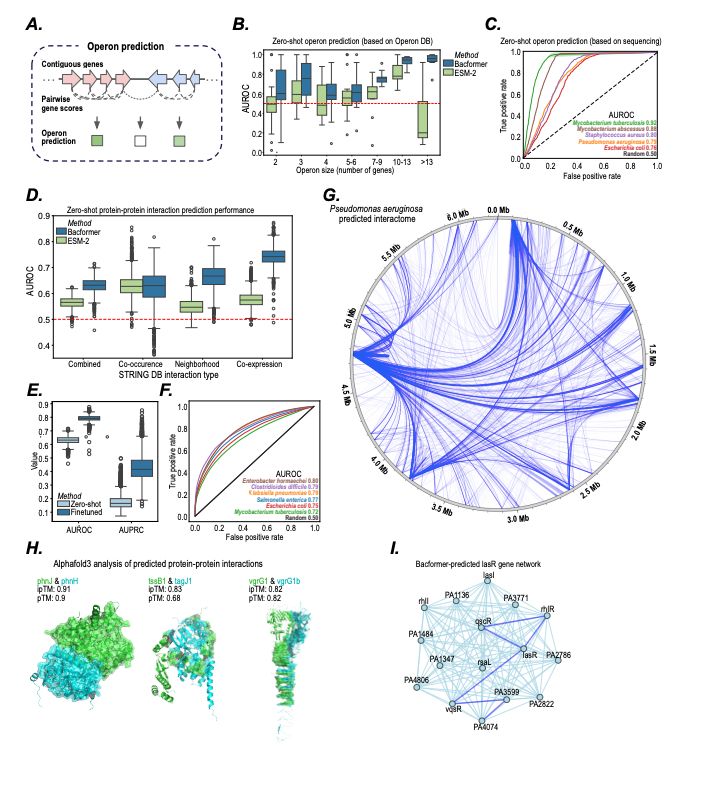
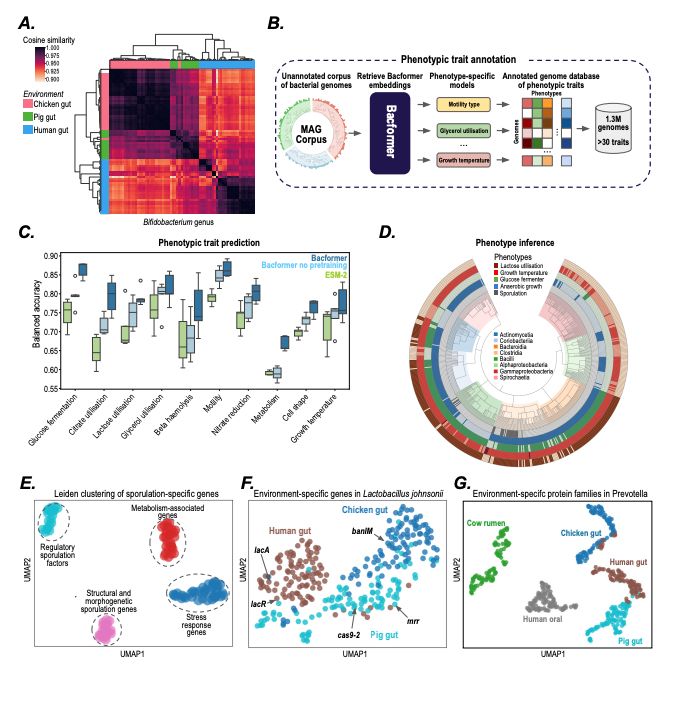
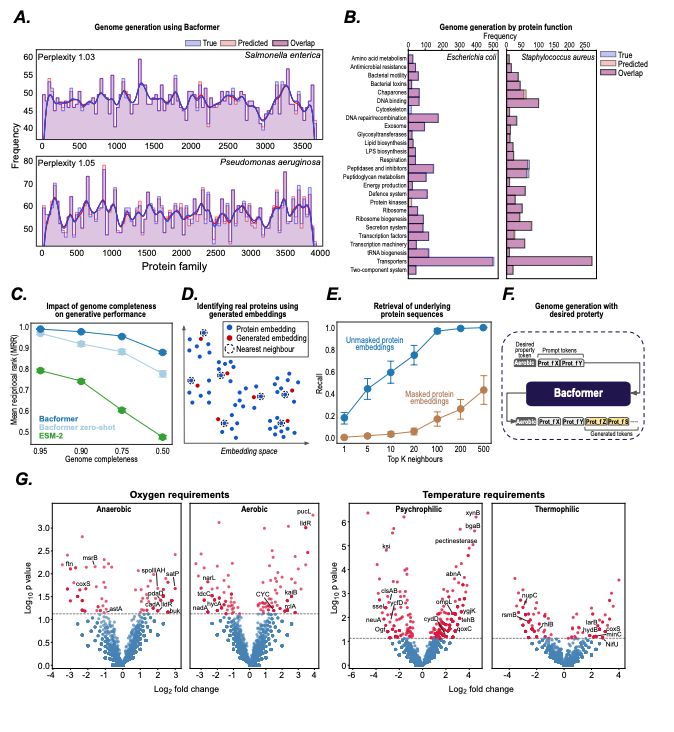
- predict protein-protein interactions, operon structure, and protein function
- infer phenotypic traits
- design synthetic genomes with desired properties
@macwiatrak.bsky.social @mariabrbic.bsky.social
@andresfloto.bsky.social

ismb-schedule.luispedro.org
ismb-schedule.luispedro.org

Check it out and share widely: www.abacbs.org/abacbs2025
Registrations and abstract submissions open next week, with abstracts due in August!

Check it out and share widely: www.abacbs.org/abacbs2025
Registrations and abstract submissions open next week, with abstracts due in August!
We predict that there are 10 archaeal and 145 bacterial novel phyla across all habitats.
#MicrobialDarkMatter #TreeOfLife #Microbiome #Archaea #Bacteria #Metagenomics
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
We predict that there are 10 archaeal and 145 bacterial novel phyla across all habitats.
#MicrobialDarkMatter #TreeOfLife #Microbiome #Archaea #Bacteria #Metagenomics
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
We’re thrilled to share our latest study tackling one of the biggest question..
"How many prokaryotic species, genera, and deeper lineages actually exist on Earth?"
Led by
@vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social , we asked a question (almost) as old as microbiology: how many prokaryotic species exist on Earth? More specifically, how much diversity is "hiding" in existing metagenomic data?
A 🧵.
We’re thrilled to share our latest study tackling one of the biggest question..
"How many prokaryotic species, genera, and deeper lineages actually exist on Earth?"
And of course to the lead authors @vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social
And of course to the lead authors @vishnuprasoodanan.bsky.social & @omaistrenko.bsky.social


