
#teamTomo
If you feel like trying one of our modules or even the full pipeline, please let us know how it goes. We are happy for any feedback and would love to improve MemBrain v2 even further to make it as helpful for the community as possible.
🧵(6/6)
If you feel like trying one of our modules or even the full pipeline, please let us know how it goes. We are happy for any feedback and would love to improve MemBrain v2 even further to make it as helpful for the community as possible.
🧵(6/6)
We focused on making MemBrain v2 smooth to work with: MemBrain-seg works with a single command line, while MemBrain-pick enables data-efficient training. We facilitate the transition between modules with several Napari functionalities like the 3D lasso to crop areas of interest.
🧵(5/6)
We focused on making MemBrain v2 smooth to work with: MemBrain-seg works with a single command line, while MemBrain-pick enables data-efficient training. We facilitate the transition between modules with several Napari functionalities like the 3D lasso to crop areas of interest.
🧵(5/6)
This module analyzes the spatial organization of particles on membranes. It takes the outputs of MemBrain-seg and MemBrain-pick to compute metrics like particle concentrations and geodesic nearest neighbor distances.
🧵(4/6)
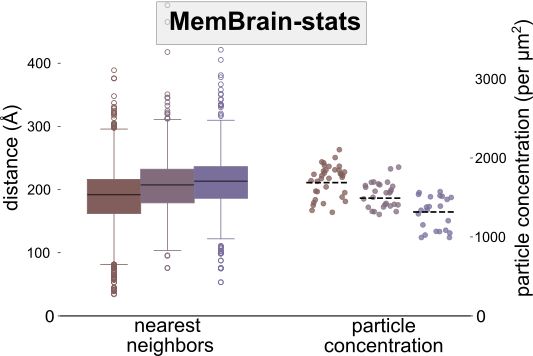
This module analyzes the spatial organization of particles on membranes. It takes the outputs of MemBrain-seg and MemBrain-pick to compute metrics like particle concentrations and geodesic nearest neighbor distances.
🧵(4/6)
If you’re interested in localizing membrane-associated particles, please give MemBrain-pick a try. It enables efficient training of a model to localize particles on membranes and works with the Surforama plugin for interactive annotation in Napari.
🔗 github.com/cellcanvas/s...
🧵(3/6)
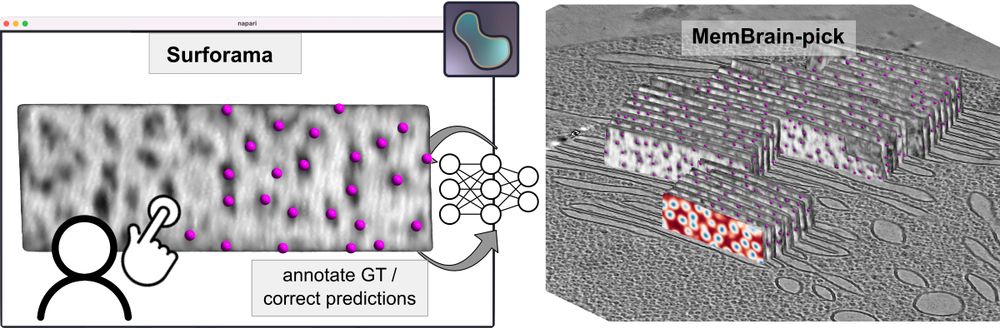
If you’re interested in localizing membrane-associated particles, please give MemBrain-pick a try. It enables efficient training of a model to localize particles on membranes and works with the Surforama plugin for interactive annotation in Napari.
🔗 github.com/cellcanvas/s...
🧵(3/6)
This module allows out-of-the-box segmentation of membranes with just a single command line.
It’s based on a U-Net architecture, trained with a diverse dataset to enable generalization to many settings.
🔗 github.com/teamtomo/mem...
🧵(2/6)
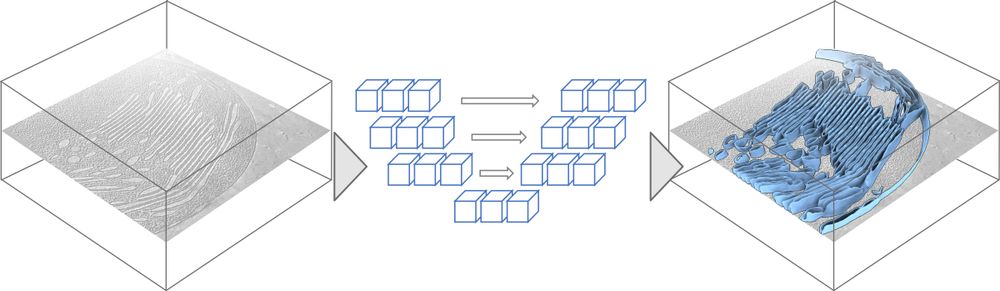
This module allows out-of-the-box segmentation of membranes with just a single command line.
It’s based on a U-Net architecture, trained with a diverse dataset to enable generalization to many settings.
🔗 github.com/teamtomo/mem...
🧵(2/6)

